
Avon-Heathcote Estuary associated circular virus 9
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.21
Get precalculated fractions of proteins
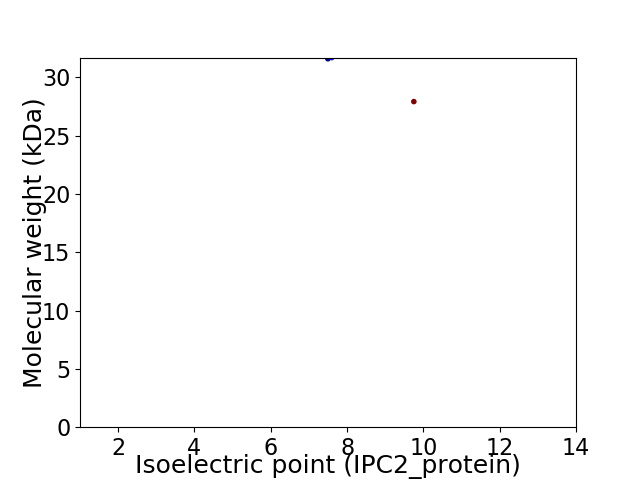
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
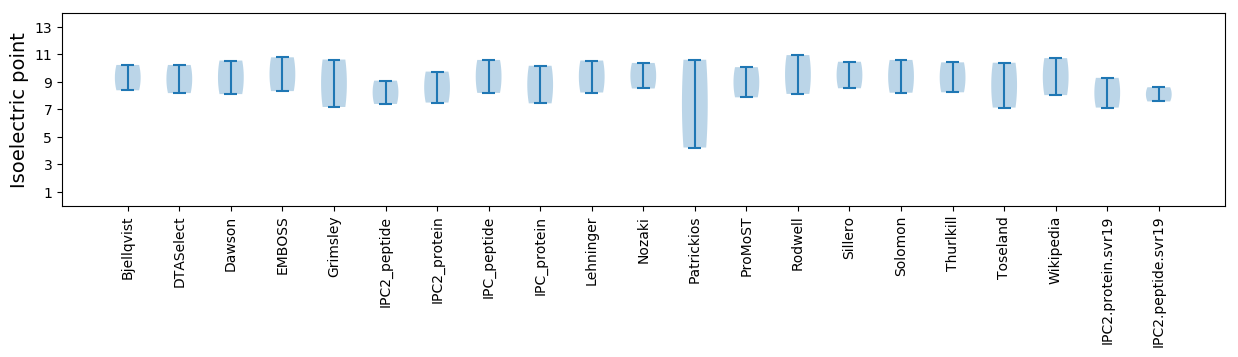
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5I9M0|A0A0C5I9M0_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 9 OX=1618260 PE=4 SV=1
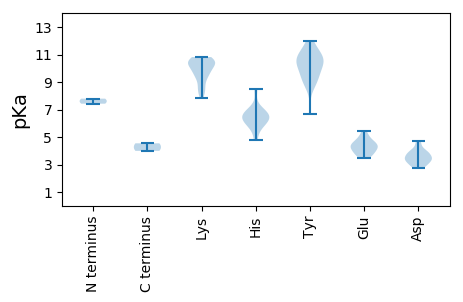
MM1 pKa = 7.43SDD3 pKa = 2.77KK4 pKa = 10.71HH5 pKa = 6.91RR6 pKa = 11.84SYY8 pKa = 11.73SFTLNNYY15 pKa = 8.83TDD17 pKa = 4.59DD18 pKa = 3.81EE19 pKa = 4.77VFALTSMCEE28 pKa = 3.93PINRR32 pKa = 11.84GVKK35 pKa = 9.5YY36 pKa = 10.47LIFGKK41 pKa = 10.28EE42 pKa = 3.68IAPTTGTPHH51 pKa = 6.38LQGCIIFTSPRR62 pKa = 11.84SFKK65 pKa = 10.68ALKK68 pKa = 10.31KK69 pKa = 9.68VVPFNRR75 pKa = 11.84ANYY78 pKa = 9.35KK79 pKa = 8.72PTISEE84 pKa = 4.18PGSARR89 pKa = 11.84YY90 pKa = 8.81CKK92 pKa = 10.08KK93 pKa = 10.77DD94 pKa = 3.01GDD96 pKa = 4.02VYY98 pKa = 11.17EE99 pKa = 4.81YY100 pKa = 8.71GTPPKK105 pKa = 9.82QGKK108 pKa = 9.82RR109 pKa = 11.84SDD111 pKa = 3.44IEE113 pKa = 3.98NVRR116 pKa = 11.84DD117 pKa = 3.45ILTSGGNIRR126 pKa = 11.84DD127 pKa = 3.92VVSTATSIQSVRR139 pKa = 11.84MAEE142 pKa = 3.52IHH144 pKa = 6.58LKK146 pKa = 10.43YY147 pKa = 10.47FEE149 pKa = 5.16KK150 pKa = 10.46KK151 pKa = 10.26RR152 pKa = 11.84NWKK155 pKa = 8.23PTVRR159 pKa = 11.84WYY161 pKa = 11.09YY162 pKa = 10.89GATGTGKK169 pKa = 10.54SKK171 pKa = 9.23TAYY174 pKa = 9.51EE175 pKa = 4.39EE176 pKa = 4.47CDD178 pKa = 3.14EE179 pKa = 4.58HH180 pKa = 8.5DD181 pKa = 4.74PYY183 pKa = 11.96VAMSTGKK190 pKa = 9.42WFEE193 pKa = 4.36GYY195 pKa = 9.4DD196 pKa = 3.31AHH198 pKa = 5.75THH200 pKa = 6.15VIIDD204 pKa = 3.88DD205 pKa = 3.62MRR207 pKa = 11.84KK208 pKa = 10.42DD209 pKa = 3.52FMKK212 pKa = 10.43FHH214 pKa = 6.98EE215 pKa = 5.0LLRR218 pKa = 11.84MLDD221 pKa = 3.49RR222 pKa = 11.84YY223 pKa = 11.27AFMVEE228 pKa = 4.26CKK230 pKa = 10.41GGSRR234 pKa = 11.84QFLATDD240 pKa = 3.82IYY242 pKa = 10.24ITSCYY247 pKa = 10.54APDD250 pKa = 3.28EE251 pKa = 4.3MFEE254 pKa = 4.09SRR256 pKa = 11.84EE257 pKa = 4.1DD258 pKa = 3.51VNQLIRR264 pKa = 11.84RR265 pKa = 11.84LDD267 pKa = 3.96EE268 pKa = 3.71IRR270 pKa = 11.84KK271 pKa = 9.53FEE273 pKa = 3.99
MM1 pKa = 7.43SDD3 pKa = 2.77KK4 pKa = 10.71HH5 pKa = 6.91RR6 pKa = 11.84SYY8 pKa = 11.73SFTLNNYY15 pKa = 8.83TDD17 pKa = 4.59DD18 pKa = 3.81EE19 pKa = 4.77VFALTSMCEE28 pKa = 3.93PINRR32 pKa = 11.84GVKK35 pKa = 9.5YY36 pKa = 10.47LIFGKK41 pKa = 10.28EE42 pKa = 3.68IAPTTGTPHH51 pKa = 6.38LQGCIIFTSPRR62 pKa = 11.84SFKK65 pKa = 10.68ALKK68 pKa = 10.31KK69 pKa = 9.68VVPFNRR75 pKa = 11.84ANYY78 pKa = 9.35KK79 pKa = 8.72PTISEE84 pKa = 4.18PGSARR89 pKa = 11.84YY90 pKa = 8.81CKK92 pKa = 10.08KK93 pKa = 10.77DD94 pKa = 3.01GDD96 pKa = 4.02VYY98 pKa = 11.17EE99 pKa = 4.81YY100 pKa = 8.71GTPPKK105 pKa = 9.82QGKK108 pKa = 9.82RR109 pKa = 11.84SDD111 pKa = 3.44IEE113 pKa = 3.98NVRR116 pKa = 11.84DD117 pKa = 3.45ILTSGGNIRR126 pKa = 11.84DD127 pKa = 3.92VVSTATSIQSVRR139 pKa = 11.84MAEE142 pKa = 3.52IHH144 pKa = 6.58LKK146 pKa = 10.43YY147 pKa = 10.47FEE149 pKa = 5.16KK150 pKa = 10.46KK151 pKa = 10.26RR152 pKa = 11.84NWKK155 pKa = 8.23PTVRR159 pKa = 11.84WYY161 pKa = 11.09YY162 pKa = 10.89GATGTGKK169 pKa = 10.54SKK171 pKa = 9.23TAYY174 pKa = 9.51EE175 pKa = 4.39EE176 pKa = 4.47CDD178 pKa = 3.14EE179 pKa = 4.58HH180 pKa = 8.5DD181 pKa = 4.74PYY183 pKa = 11.96VAMSTGKK190 pKa = 9.42WFEE193 pKa = 4.36GYY195 pKa = 9.4DD196 pKa = 3.31AHH198 pKa = 5.75THH200 pKa = 6.15VIIDD204 pKa = 3.88DD205 pKa = 3.62MRR207 pKa = 11.84KK208 pKa = 10.42DD209 pKa = 3.52FMKK212 pKa = 10.43FHH214 pKa = 6.98EE215 pKa = 5.0LLRR218 pKa = 11.84MLDD221 pKa = 3.49RR222 pKa = 11.84YY223 pKa = 11.27AFMVEE228 pKa = 4.26CKK230 pKa = 10.41GGSRR234 pKa = 11.84QFLATDD240 pKa = 3.82IYY242 pKa = 10.24ITSCYY247 pKa = 10.54APDD250 pKa = 3.28EE251 pKa = 4.3MFEE254 pKa = 4.09SRR256 pKa = 11.84EE257 pKa = 4.1DD258 pKa = 3.51VNQLIRR264 pKa = 11.84RR265 pKa = 11.84LDD267 pKa = 3.96EE268 pKa = 3.71IRR270 pKa = 11.84KK271 pKa = 9.53FEE273 pKa = 3.99
Molecular weight: 31.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5I9M0|A0A0C5I9M0_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 9 OX=1618260 PE=4 SV=1
MM1 pKa = 7.79PYY3 pKa = 9.97VRR5 pKa = 11.84KK6 pKa = 9.14PRR8 pKa = 11.84STMRR12 pKa = 11.84RR13 pKa = 11.84PYY15 pKa = 10.39KK16 pKa = 10.09KK17 pKa = 8.33RR18 pKa = 11.84TMRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 7.82PTASSMYY30 pKa = 9.68NIARR34 pKa = 11.84KK35 pKa = 8.36VLRR38 pKa = 11.84SNVEE42 pKa = 3.85TKK44 pKa = 10.79SRR46 pKa = 11.84ITSYY50 pKa = 9.38STSGMILSHH59 pKa = 5.76NVPRR63 pKa = 11.84LVASQLLQTSQGTDD77 pKa = 2.8SGATSGNRR85 pKa = 11.84VGISVQPVGIKK96 pKa = 10.29LYY98 pKa = 10.53IEE100 pKa = 4.55INQEE104 pKa = 3.51QPAAATGLLNGDD116 pKa = 2.92IWVKK120 pKa = 9.68FWVCSTHH127 pKa = 6.21HH128 pKa = 6.72SNVNTSNDD136 pKa = 3.44FLRR139 pKa = 11.84LISSNTMLAPVQRR152 pKa = 11.84RR153 pKa = 11.84THH155 pKa = 4.82NVIRR159 pKa = 11.84SFKK162 pKa = 10.68LNLRR166 pKa = 11.84NYY168 pKa = 6.65FTHH171 pKa = 6.62WNAGSAIDD179 pKa = 3.81ATPAFKK185 pKa = 10.23TRR187 pKa = 11.84TIWIPMNKK195 pKa = 8.69IKK197 pKa = 10.58QYY199 pKa = 10.85LYY201 pKa = 11.03EE202 pKa = 5.43NDD204 pKa = 3.21VTDD207 pKa = 3.26NGKK210 pKa = 9.66YY211 pKa = 10.18FDD213 pKa = 4.28YY214 pKa = 10.5CCHH217 pKa = 6.23AVAFSAHH224 pKa = 6.52PSVNTSTALAAVKK237 pKa = 10.55LNTEE241 pKa = 4.56FFFKK245 pKa = 10.82DD246 pKa = 2.96AA247 pKa = 4.54
MM1 pKa = 7.79PYY3 pKa = 9.97VRR5 pKa = 11.84KK6 pKa = 9.14PRR8 pKa = 11.84STMRR12 pKa = 11.84RR13 pKa = 11.84PYY15 pKa = 10.39KK16 pKa = 10.09KK17 pKa = 8.33RR18 pKa = 11.84TMRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 7.82PTASSMYY30 pKa = 9.68NIARR34 pKa = 11.84KK35 pKa = 8.36VLRR38 pKa = 11.84SNVEE42 pKa = 3.85TKK44 pKa = 10.79SRR46 pKa = 11.84ITSYY50 pKa = 9.38STSGMILSHH59 pKa = 5.76NVPRR63 pKa = 11.84LVASQLLQTSQGTDD77 pKa = 2.8SGATSGNRR85 pKa = 11.84VGISVQPVGIKK96 pKa = 10.29LYY98 pKa = 10.53IEE100 pKa = 4.55INQEE104 pKa = 3.51QPAAATGLLNGDD116 pKa = 2.92IWVKK120 pKa = 9.68FWVCSTHH127 pKa = 6.21HH128 pKa = 6.72SNVNTSNDD136 pKa = 3.44FLRR139 pKa = 11.84LISSNTMLAPVQRR152 pKa = 11.84RR153 pKa = 11.84THH155 pKa = 4.82NVIRR159 pKa = 11.84SFKK162 pKa = 10.68LNLRR166 pKa = 11.84NYY168 pKa = 6.65FTHH171 pKa = 6.62WNAGSAIDD179 pKa = 3.81ATPAFKK185 pKa = 10.23TRR187 pKa = 11.84TIWIPMNKK195 pKa = 8.69IKK197 pKa = 10.58QYY199 pKa = 10.85LYY201 pKa = 11.03EE202 pKa = 5.43NDD204 pKa = 3.21VTDD207 pKa = 3.26NGKK210 pKa = 9.66YY211 pKa = 10.18FDD213 pKa = 4.28YY214 pKa = 10.5CCHH217 pKa = 6.23AVAFSAHH224 pKa = 6.52PSVNTSTALAAVKK237 pKa = 10.55LNTEE241 pKa = 4.56FFFKK245 pKa = 10.82DD246 pKa = 2.96AA247 pKa = 4.54
Molecular weight: 27.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
520 |
247 |
273 |
260.0 |
29.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.346 ± 0.885 | 1.731 ± 0.34 |
5.192 ± 1.285 | 4.615 ± 1.704 |
4.615 ± 0.373 | 5.192 ± 0.752 |
2.692 ± 0.093 | 5.962 ± 0.193 |
7.115 ± 0.686 | 5.577 ± 0.592 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.077 ± 0.16 | 5.385 ± 1.518 |
4.423 ± 0.02 | 2.5 ± 0.486 |
7.115 ± 0.113 | 8.077 ± 1.078 |
8.077 ± 0.546 | 5.962 ± 0.606 |
1.346 ± 0.18 | 5.0 ± 0.626 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
