
Aliifodinibius sp. WN023
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; Balneolia; Balneolales; Balneolaceae; Aliifodinibius; unclassified Aliifodinibius
Average proteome isoelectric point is 5.69
Get precalculated fractions of proteins
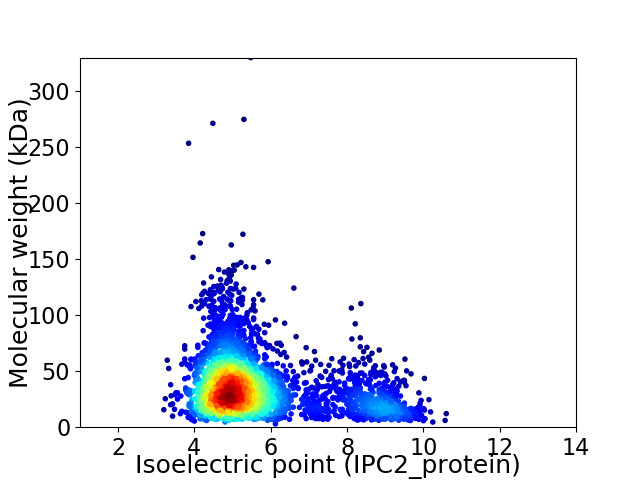
Virtual 2D-PAGE plot for 3115 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A2G9N8|A0A2A2G9N8_9BACT Succinate-semialdehyde dehydrogenase OS=Aliifodinibius sp. WN023 OX=2032627 GN=CK503_10455 PE=4 SV=1
MM1 pKa = 7.77KK2 pKa = 10.48SLLKK6 pKa = 10.08ILAMASLALLIAQCEE21 pKa = 4.55VIDD24 pKa = 4.68SNLLEE29 pKa = 5.05DD30 pKa = 4.99PNNPSPEE37 pKa = 4.02DD38 pKa = 3.64VNVDD42 pKa = 3.78FLFNNIQQEE51 pKa = 3.95ARR53 pKa = 11.84NVYY56 pKa = 10.43AGAADD61 pKa = 3.65EE62 pKa = 4.51AGEE65 pKa = 4.24MSRR68 pKa = 11.84MTYY71 pKa = 9.7MFGATYY77 pKa = 11.02ADD79 pKa = 4.5AYY81 pKa = 10.03TPTTFDD87 pKa = 3.11GLYY90 pKa = 9.87SNAYY94 pKa = 9.01ADD96 pKa = 4.5LFIDD100 pKa = 3.97VEE102 pKa = 4.2NLIPIAEE109 pKa = 4.11EE110 pKa = 3.74RR111 pKa = 11.84DD112 pKa = 3.64LTFHH116 pKa = 6.42TGAAKK121 pKa = 10.25VLKK124 pKa = 9.46AYY126 pKa = 10.77AMITLVDD133 pKa = 3.73VYY135 pKa = 11.5GDD137 pKa = 3.78VPLSNALDD145 pKa = 3.46QTNFNPEE152 pKa = 4.14LDD154 pKa = 3.99GGAAVYY160 pKa = 10.59DD161 pKa = 3.92SALVLLDD168 pKa = 4.47DD169 pKa = 6.35AIADD173 pKa = 4.18LEE175 pKa = 4.54TEE177 pKa = 4.09PTVYY181 pKa = 10.2PDD183 pKa = 3.04NDD185 pKa = 3.43LYY187 pKa = 11.48YY188 pKa = 11.14GSFEE192 pKa = 4.4DD193 pKa = 6.31RR194 pKa = 11.84EE195 pKa = 4.4DD196 pKa = 5.0QVDD199 pKa = 3.08AWIRR203 pKa = 11.84AANTIKK209 pKa = 10.76LKK211 pKa = 10.77AYY213 pKa = 10.48LNTDD217 pKa = 3.17NASAIQSLVDD227 pKa = 3.48EE228 pKa = 5.17GMLITTSTNNFTYY241 pKa = 10.64QFGTNSTNPDD251 pKa = 2.81SRR253 pKa = 11.84HH254 pKa = 5.71PSFSNNYY261 pKa = 8.48VDD263 pKa = 4.0VANDD267 pKa = 3.59YY268 pKa = 11.02MSVNYY273 pKa = 10.38LNMMLNDD280 pKa = 4.46KK281 pKa = 11.12GDD283 pKa = 3.53VDD285 pKa = 4.29PRR287 pKa = 11.84TPYY290 pKa = 10.25YY291 pKa = 10.38FYY293 pKa = 10.82RR294 pKa = 11.84QTTSDD299 pKa = 3.63PTDD302 pKa = 3.25VNLNTCISAFKK313 pKa = 10.2PNHH316 pKa = 5.77FQQNDD321 pKa = 3.83PFCLLGNGYY330 pKa = 9.07WGRR333 pKa = 11.84DD334 pKa = 3.36HH335 pKa = 7.27LIDD338 pKa = 5.9DD339 pKa = 6.24GIPPDD344 pKa = 3.89GDD346 pKa = 3.38LRR348 pKa = 11.84TTFGVYY354 pKa = 9.62PVGGAFDD361 pKa = 3.73TGQEE365 pKa = 4.04EE366 pKa = 4.82GTEE369 pKa = 3.98EE370 pKa = 5.05DD371 pKa = 3.88MGLQGAGFDD380 pKa = 4.75PILMSSFTHH389 pKa = 6.84FMLAEE394 pKa = 3.71YY395 pKa = 9.93EE396 pKa = 4.06YY397 pKa = 10.9RR398 pKa = 11.84LNGNAGAARR407 pKa = 11.84NHH409 pKa = 6.9LEE411 pKa = 3.88TAMTQSLEE419 pKa = 4.22TVRR422 pKa = 11.84ASAGTLDD429 pKa = 5.14DD430 pKa = 5.43DD431 pKa = 4.59EE432 pKa = 4.91QEE434 pKa = 4.19EE435 pKa = 4.34MSDD438 pKa = 3.65NDD440 pKa = 2.91ITNYY444 pKa = 9.53VDD446 pKa = 3.88NVLNNRR452 pKa = 11.84YY453 pKa = 9.9DD454 pKa = 3.47DD455 pKa = 3.96AGRR458 pKa = 11.84DD459 pKa = 3.72EE460 pKa = 4.95LRR462 pKa = 11.84VISKK466 pKa = 9.7EE467 pKa = 3.88FYY469 pKa = 10.34LALWTNGLEE478 pKa = 4.62AYY480 pKa = 11.33NMMRR484 pKa = 11.84RR485 pKa = 11.84TGYY488 pKa = 9.64PNKK491 pKa = 9.84QDD493 pKa = 3.44NLQPARR499 pKa = 11.84SPNPGSWYY507 pKa = 10.82NSFLYY512 pKa = 8.83PASMVEE518 pKa = 4.32RR519 pKa = 11.84NNSVSQKK526 pKa = 10.46DD527 pKa = 3.37RR528 pKa = 11.84SEE530 pKa = 4.19TVFWDD535 pKa = 3.49DD536 pKa = 2.72GTYY539 pKa = 11.15NFDD542 pKa = 3.36FF543 pKa = 5.06
MM1 pKa = 7.77KK2 pKa = 10.48SLLKK6 pKa = 10.08ILAMASLALLIAQCEE21 pKa = 4.55VIDD24 pKa = 4.68SNLLEE29 pKa = 5.05DD30 pKa = 4.99PNNPSPEE37 pKa = 4.02DD38 pKa = 3.64VNVDD42 pKa = 3.78FLFNNIQQEE51 pKa = 3.95ARR53 pKa = 11.84NVYY56 pKa = 10.43AGAADD61 pKa = 3.65EE62 pKa = 4.51AGEE65 pKa = 4.24MSRR68 pKa = 11.84MTYY71 pKa = 9.7MFGATYY77 pKa = 11.02ADD79 pKa = 4.5AYY81 pKa = 10.03TPTTFDD87 pKa = 3.11GLYY90 pKa = 9.87SNAYY94 pKa = 9.01ADD96 pKa = 4.5LFIDD100 pKa = 3.97VEE102 pKa = 4.2NLIPIAEE109 pKa = 4.11EE110 pKa = 3.74RR111 pKa = 11.84DD112 pKa = 3.64LTFHH116 pKa = 6.42TGAAKK121 pKa = 10.25VLKK124 pKa = 9.46AYY126 pKa = 10.77AMITLVDD133 pKa = 3.73VYY135 pKa = 11.5GDD137 pKa = 3.78VPLSNALDD145 pKa = 3.46QTNFNPEE152 pKa = 4.14LDD154 pKa = 3.99GGAAVYY160 pKa = 10.59DD161 pKa = 3.92SALVLLDD168 pKa = 4.47DD169 pKa = 6.35AIADD173 pKa = 4.18LEE175 pKa = 4.54TEE177 pKa = 4.09PTVYY181 pKa = 10.2PDD183 pKa = 3.04NDD185 pKa = 3.43LYY187 pKa = 11.48YY188 pKa = 11.14GSFEE192 pKa = 4.4DD193 pKa = 6.31RR194 pKa = 11.84EE195 pKa = 4.4DD196 pKa = 5.0QVDD199 pKa = 3.08AWIRR203 pKa = 11.84AANTIKK209 pKa = 10.76LKK211 pKa = 10.77AYY213 pKa = 10.48LNTDD217 pKa = 3.17NASAIQSLVDD227 pKa = 3.48EE228 pKa = 5.17GMLITTSTNNFTYY241 pKa = 10.64QFGTNSTNPDD251 pKa = 2.81SRR253 pKa = 11.84HH254 pKa = 5.71PSFSNNYY261 pKa = 8.48VDD263 pKa = 4.0VANDD267 pKa = 3.59YY268 pKa = 11.02MSVNYY273 pKa = 10.38LNMMLNDD280 pKa = 4.46KK281 pKa = 11.12GDD283 pKa = 3.53VDD285 pKa = 4.29PRR287 pKa = 11.84TPYY290 pKa = 10.25YY291 pKa = 10.38FYY293 pKa = 10.82RR294 pKa = 11.84QTTSDD299 pKa = 3.63PTDD302 pKa = 3.25VNLNTCISAFKK313 pKa = 10.2PNHH316 pKa = 5.77FQQNDD321 pKa = 3.83PFCLLGNGYY330 pKa = 9.07WGRR333 pKa = 11.84DD334 pKa = 3.36HH335 pKa = 7.27LIDD338 pKa = 5.9DD339 pKa = 6.24GIPPDD344 pKa = 3.89GDD346 pKa = 3.38LRR348 pKa = 11.84TTFGVYY354 pKa = 9.62PVGGAFDD361 pKa = 3.73TGQEE365 pKa = 4.04EE366 pKa = 4.82GTEE369 pKa = 3.98EE370 pKa = 5.05DD371 pKa = 3.88MGLQGAGFDD380 pKa = 4.75PILMSSFTHH389 pKa = 6.84FMLAEE394 pKa = 3.71YY395 pKa = 9.93EE396 pKa = 4.06YY397 pKa = 10.9RR398 pKa = 11.84LNGNAGAARR407 pKa = 11.84NHH409 pKa = 6.9LEE411 pKa = 3.88TAMTQSLEE419 pKa = 4.22TVRR422 pKa = 11.84ASAGTLDD429 pKa = 5.14DD430 pKa = 5.43DD431 pKa = 4.59EE432 pKa = 4.91QEE434 pKa = 4.19EE435 pKa = 4.34MSDD438 pKa = 3.65NDD440 pKa = 2.91ITNYY444 pKa = 9.53VDD446 pKa = 3.88NVLNNRR452 pKa = 11.84YY453 pKa = 9.9DD454 pKa = 3.47DD455 pKa = 3.96AGRR458 pKa = 11.84DD459 pKa = 3.72EE460 pKa = 4.95LRR462 pKa = 11.84VISKK466 pKa = 9.7EE467 pKa = 3.88FYY469 pKa = 10.34LALWTNGLEE478 pKa = 4.62AYY480 pKa = 11.33NMMRR484 pKa = 11.84RR485 pKa = 11.84TGYY488 pKa = 9.64PNKK491 pKa = 9.84QDD493 pKa = 3.44NLQPARR499 pKa = 11.84SPNPGSWYY507 pKa = 10.82NSFLYY512 pKa = 8.83PASMVEE518 pKa = 4.32RR519 pKa = 11.84NNSVSQKK526 pKa = 10.46DD527 pKa = 3.37RR528 pKa = 11.84SEE530 pKa = 4.19TVFWDD535 pKa = 3.49DD536 pKa = 2.72GTYY539 pKa = 11.15NFDD542 pKa = 3.36FF543 pKa = 5.06
Molecular weight: 60.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A2G817|A0A2A2G817_9BACT SAM-dependent methyltransferase OS=Aliifodinibius sp. WN023 OX=2032627 GN=CK503_13805 PE=4 SV=1
MM1 pKa = 7.78QIRR4 pKa = 11.84QPILYY9 pKa = 9.28GLVFILVLILVLVAIGGGLFNQSARR34 pKa = 11.84WPVQWQHH41 pKa = 5.3QAFNSLCHH49 pKa = 5.86QMANRR54 pKa = 11.84SFWINGQPMAVCSRR68 pKa = 11.84CIGVYY73 pKa = 10.19SGFALGWMLLPMLSLIRR90 pKa = 11.84ITRR93 pKa = 11.84LAHH96 pKa = 5.34IKK98 pKa = 10.11KK99 pKa = 9.17VLVVILLFNFIDD111 pKa = 3.81AAGNLLGLWQNTLFSRR127 pKa = 11.84AILGGMLGSSAALIFVGDD145 pKa = 4.51FFHH148 pKa = 7.66HH149 pKa = 6.14NN150 pKa = 3.3
MM1 pKa = 7.78QIRR4 pKa = 11.84QPILYY9 pKa = 9.28GLVFILVLILVLVAIGGGLFNQSARR34 pKa = 11.84WPVQWQHH41 pKa = 5.3QAFNSLCHH49 pKa = 5.86QMANRR54 pKa = 11.84SFWINGQPMAVCSRR68 pKa = 11.84CIGVYY73 pKa = 10.19SGFALGWMLLPMLSLIRR90 pKa = 11.84ITRR93 pKa = 11.84LAHH96 pKa = 5.34IKK98 pKa = 10.11KK99 pKa = 9.17VLVVILLFNFIDD111 pKa = 3.81AAGNLLGLWQNTLFSRR127 pKa = 11.84AILGGMLGSSAALIFVGDD145 pKa = 4.51FFHH148 pKa = 7.66HH149 pKa = 6.14NN150 pKa = 3.3
Molecular weight: 16.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1052852 |
25 |
2870 |
338.0 |
38.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.63 ± 0.042 | 0.607 ± 0.011 |
6.369 ± 0.04 | 7.697 ± 0.055 |
4.503 ± 0.034 | 6.898 ± 0.035 |
1.959 ± 0.02 | 7.092 ± 0.047 |
5.991 ± 0.052 | 9.159 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.41 ± 0.023 | 5.028 ± 0.037 |
3.81 ± 0.022 | 4.098 ± 0.032 |
4.373 ± 0.03 | 6.7 ± 0.04 |
5.471 ± 0.032 | 6.327 ± 0.036 |
1.209 ± 0.017 | 3.667 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
