
Alternaria longipes dsRNA virus 1
Taxonomy: Viruses; Riboviria; dsRNA viruses; unclassified dsRNA viruses
Average proteome isoelectric point is 7.76
Get precalculated fractions of proteins
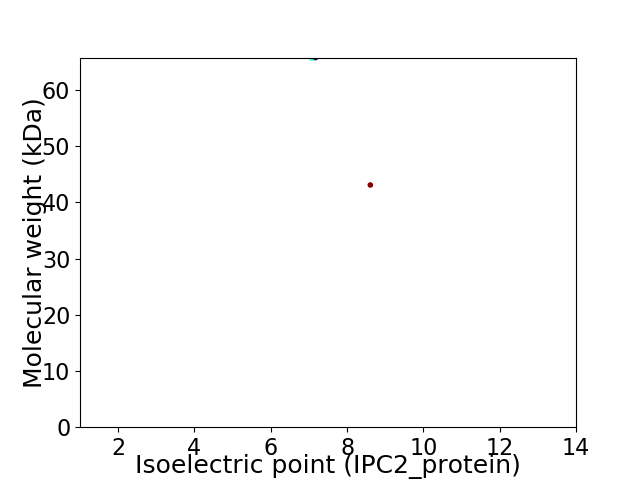
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
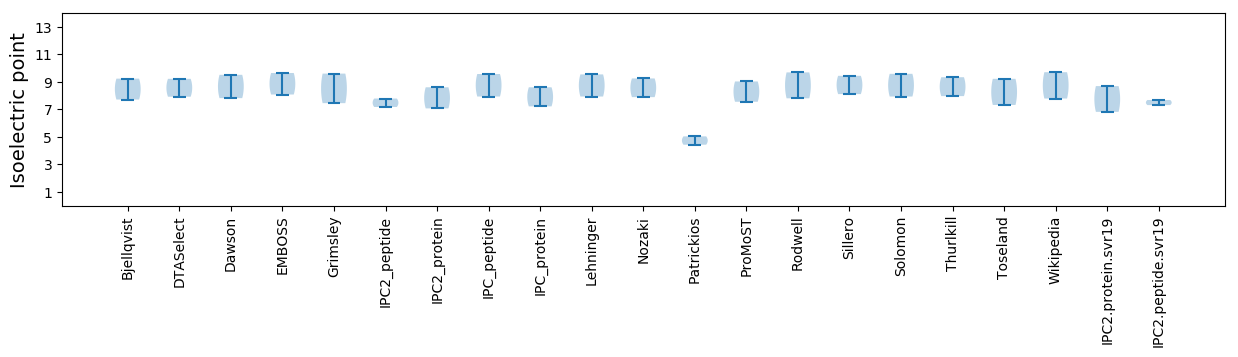
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
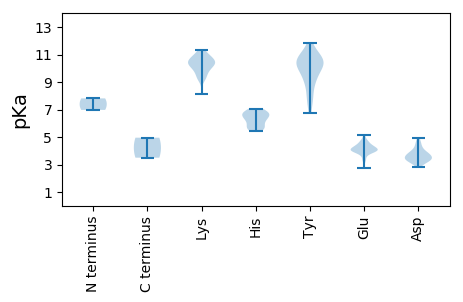
Protein with the lowest isoelectric point:
>tr|A0A076L1I6|A0A076L1I6_9VIRU Uncharacterized protein OS=Alternaria longipes dsRNA virus 1 OX=1532031 PE=4 SV=1
MM1 pKa = 7.81AEE3 pKa = 4.26NPSTTSDD10 pKa = 3.22AAGKK14 pKa = 9.84RR15 pKa = 11.84RR16 pKa = 11.84PPTEE20 pKa = 2.78WKK22 pKa = 9.43MFLEE26 pKa = 4.6NRR28 pKa = 11.84SHH30 pKa = 7.06RR31 pKa = 11.84PVPGVKK37 pKa = 10.3YY38 pKa = 9.31MGSSSHH44 pKa = 6.51LRR46 pKa = 11.84YY47 pKa = 9.99KK48 pKa = 10.39YY49 pKa = 9.79VQPEE53 pKa = 4.38TNTSSFFCKK62 pKa = 10.3FLDD65 pKa = 3.38QSTVVGGEE73 pKa = 3.79IDD75 pKa = 3.88PPSADD80 pKa = 2.79VVMSEE85 pKa = 4.18FVPPAEE91 pKa = 4.02EE92 pKa = 4.65DD93 pKa = 3.09LWKK96 pKa = 10.56HH97 pKa = 5.45VEE99 pKa = 4.69GFGQNQNRR107 pKa = 11.84GGGPSQASVSDD118 pKa = 3.18ILNEE122 pKa = 3.76LRR124 pKa = 11.84RR125 pKa = 11.84LEE127 pKa = 4.08GARR130 pKa = 11.84PIIDD134 pKa = 3.49DD135 pKa = 3.61WFEE138 pKa = 3.98ASALPKK144 pKa = 10.06IRR146 pKa = 11.84VPAHH150 pKa = 5.75TSPGIRR156 pKa = 11.84WKK158 pKa = 10.87KK159 pKa = 10.21LGYY162 pKa = 7.89KK163 pKa = 8.13TKK165 pKa = 10.46RR166 pKa = 11.84DD167 pKa = 3.65ALMPAIIEE175 pKa = 3.99ATRR178 pKa = 11.84IIGKK182 pKa = 9.67LKK184 pKa = 9.38EE185 pKa = 4.23TGHH188 pKa = 6.52EE189 pKa = 4.04YY190 pKa = 10.64DD191 pKa = 3.83VPPAGVAGRR200 pKa = 11.84GKK202 pKa = 10.08RR203 pKa = 11.84VALDD207 pKa = 3.75RR208 pKa = 11.84PDD210 pKa = 4.46DD211 pKa = 4.23PDD213 pKa = 3.6RR214 pKa = 11.84KK215 pKa = 9.19EE216 pKa = 3.66GRR218 pKa = 11.84LIVMPDD224 pKa = 3.6LVRR227 pKa = 11.84HH228 pKa = 6.16LLGAMAAAPYY238 pKa = 10.31MSMQRR243 pKa = 11.84SLDD246 pKa = 3.48KK247 pKa = 11.3SRR249 pKa = 11.84GGVMLGMGPFQEE261 pKa = 4.78SYY263 pKa = 11.2QDD265 pKa = 3.29IAGWAAGAKK274 pKa = 9.35RR275 pKa = 11.84YY276 pKa = 10.2VFIDD280 pKa = 3.94FKK282 pKa = 11.32KK283 pKa = 9.46FDD285 pKa = 3.3QRR287 pKa = 11.84IPRR290 pKa = 11.84RR291 pKa = 11.84VLRR294 pKa = 11.84AVMKK298 pKa = 10.03HH299 pKa = 5.58ISHH302 pKa = 6.84SYY304 pKa = 10.09SNGPGTGAYY313 pKa = 6.78WASEE317 pKa = 3.71FRR319 pKa = 11.84HH320 pKa = 5.63LVEE323 pKa = 5.17TEE325 pKa = 3.23IAMPNGSVYY334 pKa = 10.76KK335 pKa = 10.46KK336 pKa = 10.71FMGVASGDD344 pKa = 3.51PWTSLADD351 pKa = 3.86SYY353 pKa = 11.86ANWVMLSMACKK364 pKa = 10.33ALGLRR369 pKa = 11.84AKK371 pKa = 9.79IWTFGDD377 pKa = 3.82DD378 pKa = 3.21SVLAIDD384 pKa = 4.54EE385 pKa = 4.54GDD387 pKa = 3.4TRR389 pKa = 11.84GDD391 pKa = 3.33LLGRR395 pKa = 11.84ISEE398 pKa = 4.35WVSKK402 pKa = 10.05EE403 pKa = 3.42FGMVVSKK410 pKa = 10.49EE411 pKa = 3.92KK412 pKa = 10.94SYY414 pKa = 10.62MATDD418 pKa = 4.29LVTIDD423 pKa = 4.66EE424 pKa = 4.99DD425 pKa = 3.9PQEE428 pKa = 4.29GLSGSFLSMYY438 pKa = 10.64FLATPMGVRR447 pKa = 11.84PTRR450 pKa = 11.84PLQHH454 pKa = 6.8FYY456 pKa = 11.42EE457 pKa = 4.81LFLKK461 pKa = 10.26PEE463 pKa = 4.33RR464 pKa = 11.84NGGTVEE470 pKa = 3.95WEE472 pKa = 4.13VARR475 pKa = 11.84TSMAYY480 pKa = 10.32LVFYY484 pKa = 8.71YY485 pKa = 9.86NWKK488 pKa = 9.18VRR490 pKa = 11.84YY491 pKa = 9.0ILHH494 pKa = 6.87EE495 pKa = 3.98YY496 pKa = 9.01WDD498 pKa = 4.09WLHH501 pKa = 6.37EE502 pKa = 4.31RR503 pKa = 11.84YY504 pKa = 9.89KK505 pKa = 10.55IPEE508 pKa = 4.0LRR510 pKa = 11.84GTLDD514 pKa = 3.35DD515 pKa = 4.73LRR517 pKa = 11.84LLRR520 pKa = 11.84EE521 pKa = 4.01LDD523 pKa = 3.5IPWAHH528 pKa = 6.78FKK530 pKa = 11.03LSWLSHH536 pKa = 6.5LPPAGEE542 pKa = 4.03VEE544 pKa = 4.22LLYY547 pKa = 10.68KK548 pKa = 10.57YY549 pKa = 10.83GHH551 pKa = 5.65TCFYY555 pKa = 10.85PPVLWDD561 pKa = 3.43KK562 pKa = 11.03LYY564 pKa = 11.21SRR566 pKa = 11.84NSEE569 pKa = 4.1LPGGNSFADD578 pKa = 3.42WW579 pKa = 3.5
MM1 pKa = 7.81AEE3 pKa = 4.26NPSTTSDD10 pKa = 3.22AAGKK14 pKa = 9.84RR15 pKa = 11.84RR16 pKa = 11.84PPTEE20 pKa = 2.78WKK22 pKa = 9.43MFLEE26 pKa = 4.6NRR28 pKa = 11.84SHH30 pKa = 7.06RR31 pKa = 11.84PVPGVKK37 pKa = 10.3YY38 pKa = 9.31MGSSSHH44 pKa = 6.51LRR46 pKa = 11.84YY47 pKa = 9.99KK48 pKa = 10.39YY49 pKa = 9.79VQPEE53 pKa = 4.38TNTSSFFCKK62 pKa = 10.3FLDD65 pKa = 3.38QSTVVGGEE73 pKa = 3.79IDD75 pKa = 3.88PPSADD80 pKa = 2.79VVMSEE85 pKa = 4.18FVPPAEE91 pKa = 4.02EE92 pKa = 4.65DD93 pKa = 3.09LWKK96 pKa = 10.56HH97 pKa = 5.45VEE99 pKa = 4.69GFGQNQNRR107 pKa = 11.84GGGPSQASVSDD118 pKa = 3.18ILNEE122 pKa = 3.76LRR124 pKa = 11.84RR125 pKa = 11.84LEE127 pKa = 4.08GARR130 pKa = 11.84PIIDD134 pKa = 3.49DD135 pKa = 3.61WFEE138 pKa = 3.98ASALPKK144 pKa = 10.06IRR146 pKa = 11.84VPAHH150 pKa = 5.75TSPGIRR156 pKa = 11.84WKK158 pKa = 10.87KK159 pKa = 10.21LGYY162 pKa = 7.89KK163 pKa = 8.13TKK165 pKa = 10.46RR166 pKa = 11.84DD167 pKa = 3.65ALMPAIIEE175 pKa = 3.99ATRR178 pKa = 11.84IIGKK182 pKa = 9.67LKK184 pKa = 9.38EE185 pKa = 4.23TGHH188 pKa = 6.52EE189 pKa = 4.04YY190 pKa = 10.64DD191 pKa = 3.83VPPAGVAGRR200 pKa = 11.84GKK202 pKa = 10.08RR203 pKa = 11.84VALDD207 pKa = 3.75RR208 pKa = 11.84PDD210 pKa = 4.46DD211 pKa = 4.23PDD213 pKa = 3.6RR214 pKa = 11.84KK215 pKa = 9.19EE216 pKa = 3.66GRR218 pKa = 11.84LIVMPDD224 pKa = 3.6LVRR227 pKa = 11.84HH228 pKa = 6.16LLGAMAAAPYY238 pKa = 10.31MSMQRR243 pKa = 11.84SLDD246 pKa = 3.48KK247 pKa = 11.3SRR249 pKa = 11.84GGVMLGMGPFQEE261 pKa = 4.78SYY263 pKa = 11.2QDD265 pKa = 3.29IAGWAAGAKK274 pKa = 9.35RR275 pKa = 11.84YY276 pKa = 10.2VFIDD280 pKa = 3.94FKK282 pKa = 11.32KK283 pKa = 9.46FDD285 pKa = 3.3QRR287 pKa = 11.84IPRR290 pKa = 11.84RR291 pKa = 11.84VLRR294 pKa = 11.84AVMKK298 pKa = 10.03HH299 pKa = 5.58ISHH302 pKa = 6.84SYY304 pKa = 10.09SNGPGTGAYY313 pKa = 6.78WASEE317 pKa = 3.71FRR319 pKa = 11.84HH320 pKa = 5.63LVEE323 pKa = 5.17TEE325 pKa = 3.23IAMPNGSVYY334 pKa = 10.76KK335 pKa = 10.46KK336 pKa = 10.71FMGVASGDD344 pKa = 3.51PWTSLADD351 pKa = 3.86SYY353 pKa = 11.86ANWVMLSMACKK364 pKa = 10.33ALGLRR369 pKa = 11.84AKK371 pKa = 9.79IWTFGDD377 pKa = 3.82DD378 pKa = 3.21SVLAIDD384 pKa = 4.54EE385 pKa = 4.54GDD387 pKa = 3.4TRR389 pKa = 11.84GDD391 pKa = 3.33LLGRR395 pKa = 11.84ISEE398 pKa = 4.35WVSKK402 pKa = 10.05EE403 pKa = 3.42FGMVVSKK410 pKa = 10.49EE411 pKa = 3.92KK412 pKa = 10.94SYY414 pKa = 10.62MATDD418 pKa = 4.29LVTIDD423 pKa = 4.66EE424 pKa = 4.99DD425 pKa = 3.9PQEE428 pKa = 4.29GLSGSFLSMYY438 pKa = 10.64FLATPMGVRR447 pKa = 11.84PTRR450 pKa = 11.84PLQHH454 pKa = 6.8FYY456 pKa = 11.42EE457 pKa = 4.81LFLKK461 pKa = 10.26PEE463 pKa = 4.33RR464 pKa = 11.84NGGTVEE470 pKa = 3.95WEE472 pKa = 4.13VARR475 pKa = 11.84TSMAYY480 pKa = 10.32LVFYY484 pKa = 8.71YY485 pKa = 9.86NWKK488 pKa = 9.18VRR490 pKa = 11.84YY491 pKa = 9.0ILHH494 pKa = 6.87EE495 pKa = 3.98YY496 pKa = 9.01WDD498 pKa = 4.09WLHH501 pKa = 6.37EE502 pKa = 4.31RR503 pKa = 11.84YY504 pKa = 9.89KK505 pKa = 10.55IPEE508 pKa = 4.0LRR510 pKa = 11.84GTLDD514 pKa = 3.35DD515 pKa = 4.73LRR517 pKa = 11.84LLRR520 pKa = 11.84EE521 pKa = 4.01LDD523 pKa = 3.5IPWAHH528 pKa = 6.78FKK530 pKa = 11.03LSWLSHH536 pKa = 6.5LPPAGEE542 pKa = 4.03VEE544 pKa = 4.22LLYY547 pKa = 10.68KK548 pKa = 10.57YY549 pKa = 10.83GHH551 pKa = 5.65TCFYY555 pKa = 10.85PPVLWDD561 pKa = 3.43KK562 pKa = 11.03LYY564 pKa = 11.21SRR566 pKa = 11.84NSEE569 pKa = 4.1LPGGNSFADD578 pKa = 3.42WW579 pKa = 3.5
Molecular weight: 65.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076L1I6|A0A076L1I6_9VIRU Uncharacterized protein OS=Alternaria longipes dsRNA virus 1 OX=1532031 PE=4 SV=1
MM1 pKa = 6.97VVAEE5 pKa = 4.67ALRR8 pKa = 11.84MSLEE12 pKa = 4.29RR13 pKa = 11.84SWSSTLQWIRR23 pKa = 11.84VGTAWEE29 pKa = 3.98PRR31 pKa = 11.84PPVDD35 pKa = 4.9KK36 pKa = 11.34SMEE39 pKa = 4.4TTGPPPSGTGAPGSQTPPAGQGSARR64 pKa = 11.84PTQHH68 pKa = 6.15EE69 pKa = 4.3RR70 pKa = 11.84SASQPAANRR79 pKa = 11.84EE80 pKa = 3.92ARR82 pKa = 11.84TPPARR87 pKa = 11.84PAEE90 pKa = 4.1YY91 pKa = 10.06TEE93 pKa = 4.0LAGAFEE99 pKa = 4.41RR100 pKa = 11.84RR101 pKa = 11.84PRR103 pKa = 11.84PQTPIQPIRR112 pKa = 11.84GLNVTSPEE120 pKa = 3.73RR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84SGGSSMPPPPPPKK136 pKa = 9.75KK137 pKa = 10.43DD138 pKa = 2.96GGTAKK143 pKa = 10.2SAPPADD149 pKa = 4.0PDD151 pKa = 3.05AWALRR156 pKa = 11.84IYY158 pKa = 9.96TIVRR162 pKa = 11.84QWHH165 pKa = 6.77DD166 pKa = 3.76LNGVTGVVTDD176 pKa = 4.34PRR178 pKa = 11.84SLFPGDD184 pKa = 2.94WTLGRR189 pKa = 11.84TRR191 pKa = 11.84LVYY194 pKa = 10.68DD195 pKa = 3.71PGVAGRR201 pKa = 11.84YY202 pKa = 7.39LRR204 pKa = 11.84RR205 pKa = 11.84MLPDD209 pKa = 3.64LKK211 pKa = 10.69CYY213 pKa = 8.11TSKK216 pKa = 10.86GVEE219 pKa = 4.02TAGVVQGSDD228 pKa = 3.26DD229 pKa = 3.74VKK231 pKa = 9.78TARR234 pKa = 11.84ITAEE238 pKa = 3.37WRR240 pKa = 11.84PPVHH244 pKa = 6.58LVAEE248 pKa = 4.12ILSYY252 pKa = 10.61KK253 pKa = 9.09SEE255 pKa = 4.13RR256 pKa = 11.84TVSADD261 pKa = 3.28EE262 pKa = 4.38VVALAQSAPDD272 pKa = 3.46ALEE275 pKa = 3.93KK276 pKa = 10.82LKK278 pKa = 10.88EE279 pKa = 4.08AGKK282 pKa = 9.85SLKK285 pKa = 10.65AHH287 pKa = 6.21FTRR290 pKa = 11.84LLDD293 pKa = 3.51MVALRR298 pKa = 11.84RR299 pKa = 11.84SPAYY303 pKa = 10.31NEE305 pKa = 3.31TAARR309 pKa = 11.84YY310 pKa = 7.84KK311 pKa = 10.81AQLKK315 pKa = 10.21VLIQEE320 pKa = 4.36QYY322 pKa = 10.51NHH324 pKa = 5.84VLALKK329 pKa = 10.21QATKK333 pKa = 10.53RR334 pKa = 11.84VNDD337 pKa = 4.04LLAEE341 pKa = 4.5RR342 pKa = 11.84DD343 pKa = 3.69AEE345 pKa = 4.63LARR348 pKa = 11.84LDD350 pKa = 4.1PGYY353 pKa = 9.26TPKK356 pKa = 10.6KK357 pKa = 10.33QSAQDD362 pKa = 3.18ALARR366 pKa = 11.84YY367 pKa = 9.16GIDD370 pKa = 3.28LGSEE374 pKa = 4.07GGEE377 pKa = 4.11EE378 pKa = 4.16SWEE381 pKa = 4.01EE382 pKa = 3.91ANPDD386 pKa = 3.51NPLPNLDD393 pKa = 3.75FF394 pKa = 4.95
MM1 pKa = 6.97VVAEE5 pKa = 4.67ALRR8 pKa = 11.84MSLEE12 pKa = 4.29RR13 pKa = 11.84SWSSTLQWIRR23 pKa = 11.84VGTAWEE29 pKa = 3.98PRR31 pKa = 11.84PPVDD35 pKa = 4.9KK36 pKa = 11.34SMEE39 pKa = 4.4TTGPPPSGTGAPGSQTPPAGQGSARR64 pKa = 11.84PTQHH68 pKa = 6.15EE69 pKa = 4.3RR70 pKa = 11.84SASQPAANRR79 pKa = 11.84EE80 pKa = 3.92ARR82 pKa = 11.84TPPARR87 pKa = 11.84PAEE90 pKa = 4.1YY91 pKa = 10.06TEE93 pKa = 4.0LAGAFEE99 pKa = 4.41RR100 pKa = 11.84RR101 pKa = 11.84PRR103 pKa = 11.84PQTPIQPIRR112 pKa = 11.84GLNVTSPEE120 pKa = 3.73RR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84SGGSSMPPPPPPKK136 pKa = 9.75KK137 pKa = 10.43DD138 pKa = 2.96GGTAKK143 pKa = 10.2SAPPADD149 pKa = 4.0PDD151 pKa = 3.05AWALRR156 pKa = 11.84IYY158 pKa = 9.96TIVRR162 pKa = 11.84QWHH165 pKa = 6.77DD166 pKa = 3.76LNGVTGVVTDD176 pKa = 4.34PRR178 pKa = 11.84SLFPGDD184 pKa = 2.94WTLGRR189 pKa = 11.84TRR191 pKa = 11.84LVYY194 pKa = 10.68DD195 pKa = 3.71PGVAGRR201 pKa = 11.84YY202 pKa = 7.39LRR204 pKa = 11.84RR205 pKa = 11.84MLPDD209 pKa = 3.64LKK211 pKa = 10.69CYY213 pKa = 8.11TSKK216 pKa = 10.86GVEE219 pKa = 4.02TAGVVQGSDD228 pKa = 3.26DD229 pKa = 3.74VKK231 pKa = 9.78TARR234 pKa = 11.84ITAEE238 pKa = 3.37WRR240 pKa = 11.84PPVHH244 pKa = 6.58LVAEE248 pKa = 4.12ILSYY252 pKa = 10.61KK253 pKa = 9.09SEE255 pKa = 4.13RR256 pKa = 11.84TVSADD261 pKa = 3.28EE262 pKa = 4.38VVALAQSAPDD272 pKa = 3.46ALEE275 pKa = 3.93KK276 pKa = 10.82LKK278 pKa = 10.88EE279 pKa = 4.08AGKK282 pKa = 9.85SLKK285 pKa = 10.65AHH287 pKa = 6.21FTRR290 pKa = 11.84LLDD293 pKa = 3.51MVALRR298 pKa = 11.84RR299 pKa = 11.84SPAYY303 pKa = 10.31NEE305 pKa = 3.31TAARR309 pKa = 11.84YY310 pKa = 7.84KK311 pKa = 10.81AQLKK315 pKa = 10.21VLIQEE320 pKa = 4.36QYY322 pKa = 10.51NHH324 pKa = 5.84VLALKK329 pKa = 10.21QATKK333 pKa = 10.53RR334 pKa = 11.84VNDD337 pKa = 4.04LLAEE341 pKa = 4.5RR342 pKa = 11.84DD343 pKa = 3.69AEE345 pKa = 4.63LARR348 pKa = 11.84LDD350 pKa = 4.1PGYY353 pKa = 9.26TPKK356 pKa = 10.6KK357 pKa = 10.33QSAQDD362 pKa = 3.18ALARR366 pKa = 11.84YY367 pKa = 9.16GIDD370 pKa = 3.28LGSEE374 pKa = 4.07GGEE377 pKa = 4.11EE378 pKa = 4.16SWEE381 pKa = 4.01EE382 pKa = 3.91ANPDD386 pKa = 3.51NPLPNLDD393 pKa = 3.75FF394 pKa = 4.95
Molecular weight: 43.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
973 |
394 |
579 |
486.5 |
54.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.941 ± 1.513 | 0.411 ± 0.096 |
5.755 ± 0.26 | 6.475 ± 0.076 |
2.775 ± 1.074 | 7.811 ± 0.43 |
2.055 ± 0.48 | 3.289 ± 0.613 |
5.242 ± 0.411 | 8.736 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.775 ± 0.764 | 2.261 ± 0.014 |
8.222 ± 1.332 | 2.775 ± 0.785 |
7.708 ± 0.717 | 7.194 ± 0.208 |
5.036 ± 0.954 | 6.166 ± 0.046 |
2.672 ± 0.391 | 3.7 ± 0.554 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
