
Sulfolobales Mexican fusellovirus 1
Taxonomy: Viruses; Fuselloviridae; unclassified Fuselloviridae
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
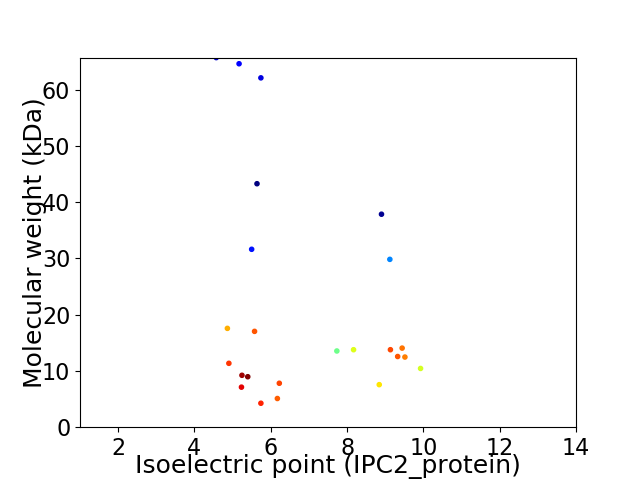
Virtual 2D-PAGE plot for 24 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M1SVC2|M1SVC2_9VIRU VP2-like protein OS=Sulfolobales Mexican fusellovirus 1 OX=1298531 GN=SMF1_0024 PE=4 SV=1
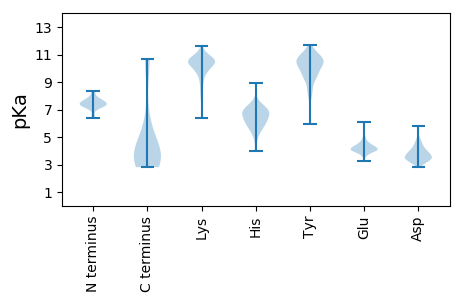
MM1 pKa = 7.39KK2 pKa = 10.37KK3 pKa = 10.27AVLVLFLLILPVLAFIPVTTNASTSGSPSITISYY37 pKa = 8.64PSTVIAGQKK46 pKa = 9.95VPITFQFNNTISSTYY61 pKa = 10.61LPILGYY67 pKa = 9.87SVSYY71 pKa = 8.16TQSGSSYY78 pKa = 10.74SYY80 pKa = 11.09TDD82 pKa = 3.49GDD84 pKa = 3.53TSGAYY89 pKa = 9.77AVTTLGANGSVIVINYY105 pKa = 7.99VGTYY109 pKa = 9.56KK110 pKa = 10.67VLEE113 pKa = 4.29GTFPGIVLYY122 pKa = 10.87GGGFEE127 pKa = 4.74TVNPSNGIATLDD139 pKa = 3.58TQQPSTATGVLVTFNGYY156 pKa = 7.96FQYY159 pKa = 11.15LSGGSWHH166 pKa = 6.38NASTSLPYY174 pKa = 10.35YY175 pKa = 10.08GSYY178 pKa = 10.11SSGYY182 pKa = 8.15IQPSSAVTWVIVFEE196 pKa = 4.62DD197 pKa = 4.03SNGEE201 pKa = 4.07TLLKK205 pKa = 10.37SISINGVTYY214 pKa = 9.13TINVLAPVPWNFTYY228 pKa = 10.89VGVRR232 pKa = 11.84PDD234 pKa = 3.45NSNDD238 pKa = 3.8GIGVSTFTVVTPSPYY253 pKa = 8.85QTYY256 pKa = 10.8DD257 pKa = 2.87VYY259 pKa = 11.65LNNNLTVTGEE269 pKa = 4.32TNMSGMGSYY278 pKa = 10.53SFTMPSSSVSVTVEE292 pKa = 3.72WVSANLNKK300 pKa = 9.84TVVLTPVTPSISITYY315 pKa = 9.14PSASYY320 pKa = 10.54AGEE323 pKa = 3.78AVKK326 pKa = 9.11ITFQFLLSNGSTTTEE341 pKa = 4.13EE342 pKa = 4.33ANQSYY347 pKa = 11.09DD348 pKa = 3.44VFIGKK353 pKa = 7.33TLNATGVTNSTGFGTFTFTMPSAPTTVTVEE383 pKa = 4.4WISANLNKK391 pKa = 9.81TVTVNLLAPQISINAPSTVYY411 pKa = 10.44AGQSVPVTFQFLILNEE427 pKa = 4.44SPTTTTAIAQANQGYY442 pKa = 9.25KK443 pKa = 10.1FYY445 pKa = 10.95LGSTLFTTGEE455 pKa = 4.38TNSTGYY461 pKa = 8.66GTVTFTMPSSAITVTVTWVSFNLNKK486 pKa = 10.37SVTLSPSSSTTTSSTTTTSSSTTSSTTSSTTTSTTSSSSTTTTSSTTSSSSTTTSSPDD544 pKa = 2.92AFQLVEE550 pKa = 4.24LTGLAPVVAIIFTFVFMGVTYY571 pKa = 10.26KK572 pKa = 10.3IAGRR576 pKa = 11.84IPAIIVAIITTVAFTFLLFLPLPYY600 pKa = 9.84TLAMLFAFIAGLIYY614 pKa = 10.27TRR616 pKa = 11.84LGGGEE621 pKa = 4.07EE622 pKa = 4.28SGDD625 pKa = 3.51
MM1 pKa = 7.39KK2 pKa = 10.37KK3 pKa = 10.27AVLVLFLLILPVLAFIPVTTNASTSGSPSITISYY37 pKa = 8.64PSTVIAGQKK46 pKa = 9.95VPITFQFNNTISSTYY61 pKa = 10.61LPILGYY67 pKa = 9.87SVSYY71 pKa = 8.16TQSGSSYY78 pKa = 10.74SYY80 pKa = 11.09TDD82 pKa = 3.49GDD84 pKa = 3.53TSGAYY89 pKa = 9.77AVTTLGANGSVIVINYY105 pKa = 7.99VGTYY109 pKa = 9.56KK110 pKa = 10.67VLEE113 pKa = 4.29GTFPGIVLYY122 pKa = 10.87GGGFEE127 pKa = 4.74TVNPSNGIATLDD139 pKa = 3.58TQQPSTATGVLVTFNGYY156 pKa = 7.96FQYY159 pKa = 11.15LSGGSWHH166 pKa = 6.38NASTSLPYY174 pKa = 10.35YY175 pKa = 10.08GSYY178 pKa = 10.11SSGYY182 pKa = 8.15IQPSSAVTWVIVFEE196 pKa = 4.62DD197 pKa = 4.03SNGEE201 pKa = 4.07TLLKK205 pKa = 10.37SISINGVTYY214 pKa = 9.13TINVLAPVPWNFTYY228 pKa = 10.89VGVRR232 pKa = 11.84PDD234 pKa = 3.45NSNDD238 pKa = 3.8GIGVSTFTVVTPSPYY253 pKa = 8.85QTYY256 pKa = 10.8DD257 pKa = 2.87VYY259 pKa = 11.65LNNNLTVTGEE269 pKa = 4.32TNMSGMGSYY278 pKa = 10.53SFTMPSSSVSVTVEE292 pKa = 3.72WVSANLNKK300 pKa = 9.84TVVLTPVTPSISITYY315 pKa = 9.14PSASYY320 pKa = 10.54AGEE323 pKa = 3.78AVKK326 pKa = 9.11ITFQFLLSNGSTTTEE341 pKa = 4.13EE342 pKa = 4.33ANQSYY347 pKa = 11.09DD348 pKa = 3.44VFIGKK353 pKa = 7.33TLNATGVTNSTGFGTFTFTMPSAPTTVTVEE383 pKa = 4.4WISANLNKK391 pKa = 9.81TVTVNLLAPQISINAPSTVYY411 pKa = 10.44AGQSVPVTFQFLILNEE427 pKa = 4.44SPTTTTAIAQANQGYY442 pKa = 9.25KK443 pKa = 10.1FYY445 pKa = 10.95LGSTLFTTGEE455 pKa = 4.38TNSTGYY461 pKa = 8.66GTVTFTMPSSAITVTVTWVSFNLNKK486 pKa = 10.37SVTLSPSSSTTTSSTTTTSSSTTSSTTSSTTTSTTSSSSTTTTSSTTSSSSTTTSSPDD544 pKa = 2.92AFQLVEE550 pKa = 4.24LTGLAPVVAIIFTFVFMGVTYY571 pKa = 10.26KK572 pKa = 10.3IAGRR576 pKa = 11.84IPAIIVAIITTVAFTFLLFLPLPYY600 pKa = 9.84TLAMLFAFIAGLIYY614 pKa = 10.27TRR616 pKa = 11.84LGGGEE621 pKa = 4.07EE622 pKa = 4.28SGDD625 pKa = 3.51
Molecular weight: 65.68 kDa
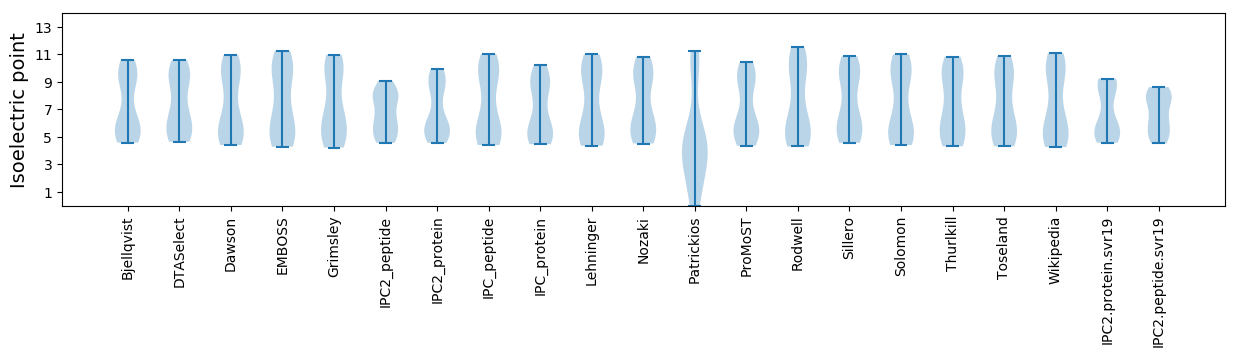
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M1T2S8|M1T2S8_9VIRU Uncharacterized protein OS=Sulfolobales Mexican fusellovirus 1 OX=1298531 GN=SMF1_0002 PE=4 SV=1
MM1 pKa = 8.2AEE3 pKa = 4.26EE4 pKa = 4.09KK5 pKa = 9.84WIQKK9 pKa = 9.14AVKK12 pKa = 9.87HH13 pKa = 6.03KK14 pKa = 10.74GRR16 pKa = 11.84VHH18 pKa = 7.16RR19 pKa = 11.84YY20 pKa = 8.46LEE22 pKa = 3.84HH23 pKa = 7.34LYY25 pKa = 10.2GHH27 pKa = 7.03KK28 pKa = 10.47AFKK31 pKa = 10.67EE32 pKa = 3.87NGDD35 pKa = 3.57IKK37 pKa = 11.15VKK39 pKa = 9.17YY40 pKa = 9.52LNMAIRR46 pKa = 11.84HH47 pKa = 4.76VKK49 pKa = 7.96RR50 pKa = 11.84AKK52 pKa = 9.85MPEE55 pKa = 3.77HH56 pKa = 6.98EE57 pKa = 4.51KK58 pKa = 10.8RR59 pKa = 11.84SLLSALYY66 pKa = 8.82LAKK69 pKa = 10.48RR70 pKa = 11.84LRR72 pKa = 11.84HH73 pKa = 4.15MHH75 pKa = 5.93KK76 pKa = 9.81HH77 pKa = 5.97RR78 pKa = 11.84KK79 pKa = 7.2HH80 pKa = 5.68HH81 pKa = 5.29HH82 pKa = 5.7HH83 pKa = 6.87KK84 pKa = 10.52KK85 pKa = 10.4
MM1 pKa = 8.2AEE3 pKa = 4.26EE4 pKa = 4.09KK5 pKa = 9.84WIQKK9 pKa = 9.14AVKK12 pKa = 9.87HH13 pKa = 6.03KK14 pKa = 10.74GRR16 pKa = 11.84VHH18 pKa = 7.16RR19 pKa = 11.84YY20 pKa = 8.46LEE22 pKa = 3.84HH23 pKa = 7.34LYY25 pKa = 10.2GHH27 pKa = 7.03KK28 pKa = 10.47AFKK31 pKa = 10.67EE32 pKa = 3.87NGDD35 pKa = 3.57IKK37 pKa = 11.15VKK39 pKa = 9.17YY40 pKa = 9.52LNMAIRR46 pKa = 11.84HH47 pKa = 4.76VKK49 pKa = 7.96RR50 pKa = 11.84AKK52 pKa = 9.85MPEE55 pKa = 3.77HH56 pKa = 6.98EE57 pKa = 4.51KK58 pKa = 10.8RR59 pKa = 11.84SLLSALYY66 pKa = 8.82LAKK69 pKa = 10.48RR70 pKa = 11.84LRR72 pKa = 11.84HH73 pKa = 4.15MHH75 pKa = 5.93KK76 pKa = 9.81HH77 pKa = 5.97RR78 pKa = 11.84KK79 pKa = 7.2HH80 pKa = 5.68HH81 pKa = 5.29HH82 pKa = 5.7HH83 pKa = 6.87KK84 pKa = 10.52KK85 pKa = 10.4
Molecular weight: 10.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4637 |
38 |
625 |
193.2 |
21.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.534 ± 0.346 | 0.518 ± 0.15 |
3.084 ± 0.446 | 7.074 ± 1.039 |
4.831 ± 0.49 | 6.211 ± 0.454 |
1.574 ± 0.419 | 6.189 ± 0.462 |
6.427 ± 1.059 | 10.567 ± 0.609 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.264 ± 0.21 | 4.119 ± 0.408 |
4.701 ± 0.428 | 2.458 ± 0.165 |
4.205 ± 0.816 | 7.16 ± 0.974 |
7.699 ± 1.334 | 8.13 ± 0.378 |
1.121 ± 0.186 | 5.133 ± 0.44 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
