
Cytophagaceae bacterium FSY-15
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; unclassified Cytophagaceae
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
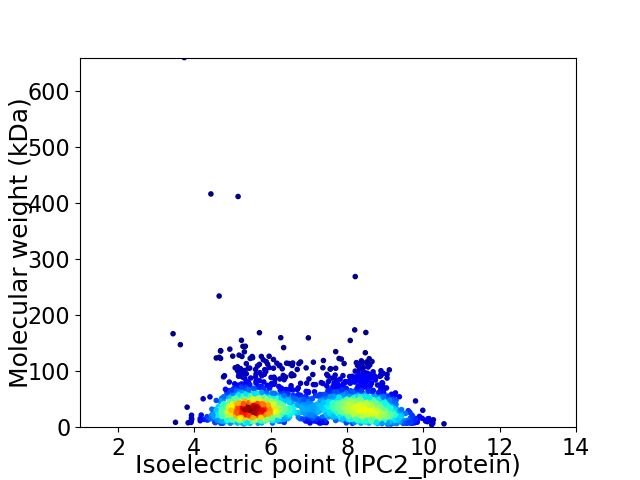
Virtual 2D-PAGE plot for 2320 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A437PW34|A0A437PW34_9BACT SDR family NAD(P)-dependent oxidoreductase OS=Cytophagaceae bacterium FSY-15 OX=1862386 GN=EOJ36_00295 PE=3 SV=1
II1 pKa = 6.87ATVKK5 pKa = 10.41SGSSVTTNVLANDD18 pKa = 3.16VCEE21 pKa = 4.6NPNCTLSNPTIVTQPAHH38 pKa = 5.49GTVTVNPDD46 pKa = 2.68GTLTYY51 pKa = 10.04TPAAGFTGTDD61 pKa = 2.91VLTYY65 pKa = 7.88QVCDD69 pKa = 3.46NSVNPAVCKK78 pKa = 9.27TATVTYY84 pKa = 8.42TVQPATAAPTTTAADD99 pKa = 3.9DD100 pKa = 4.09ASTTNSNSPVSGNVLANDD118 pKa = 4.12GSTNPNAVLTVAPQNTTIPGKK139 pKa = 9.35GTIVMNADD147 pKa = 3.14GSFTFTPTGGFVGTVDD163 pKa = 3.99IPYY166 pKa = 8.67TVCDD170 pKa = 3.47NANPANCSTATLHH183 pKa = 6.73IVVEE187 pKa = 4.17PAPTVIAPDD196 pKa = 4.1FNNGLINKK204 pKa = 8.21PIAGSLATNDD214 pKa = 3.32VVAGGGTYY222 pKa = 7.65GTPVAATSNPTGATFTVNPDD242 pKa = 2.68GSYY245 pKa = 10.93SFTGTQPGVYY255 pKa = 8.66TYY257 pKa = 10.79NVPVCPAGQTTNCPLSPVTITVVDD281 pKa = 4.1PTAANVAPAVNPDD294 pKa = 2.88IATVKK299 pKa = 10.25SGSSVTTNVLANDD312 pKa = 3.16VCEE315 pKa = 4.6NPNCTLSNPTIVTQPAHH332 pKa = 5.49GTVTVNPDD340 pKa = 2.68GTLTYY345 pKa = 10.04TPAAGFTGTDD355 pKa = 2.91VLTYY359 pKa = 7.88QVCDD363 pKa = 3.46NSVNPAVCKK372 pKa = 9.92STTVTYY378 pKa = 9.08TVQPATAAPTTTAADD393 pKa = 3.79DD394 pKa = 3.93AASTKK399 pKa = 10.79GNVPATGNVLRR410 pKa = 11.84NDD412 pKa = 3.47ASTDD416 pKa = 3.45PNAVLTVTPQSTTIPGKK433 pKa = 9.33GTIVMNADD441 pKa = 2.99GSYY444 pKa = 9.39TFTPAPGFTGTVNIPYY460 pKa = 7.87TVCDD464 pKa = 3.41NANPANCSTATLHH477 pKa = 6.47VVVEE481 pKa = 4.32PPVDD485 pKa = 3.68LVGDD489 pKa = 4.16INATNINVPVNGNVSTNDD507 pKa = 3.4SAPGSVVYY515 pKa = 7.49GTPVASANNPSGATLTMNPDD535 pKa = 2.89GTYY538 pKa = 10.71SFTATTPGKK547 pKa = 10.31YY548 pKa = 10.27VYY550 pKa = 10.12DD551 pKa = 4.01VPNCLPGQTTGCPTTPLVITVKK573 pKa = 10.85DD574 pKa = 3.91PSATNNLPVVNPDD587 pKa = 2.8IATVRR592 pKa = 11.84AGTSVTTNVLANDD605 pKa = 3.07KK606 pKa = 9.13CTNQGCSLNPASVAITAQPAHH627 pKa = 6.01GTVTVNPDD635 pKa = 2.68GTLTYY640 pKa = 10.04TPAAGFTGTDD650 pKa = 2.87VLTYY654 pKa = 9.78KK655 pKa = 10.49VCDD658 pKa = 3.29NMTPANCQTATVTYY672 pKa = 8.71TVVPANSVLTTAADD686 pKa = 3.95DD687 pKa = 4.0YY688 pKa = 10.9VTTKK692 pKa = 9.69GTGIATGNVLGNDD705 pKa = 3.31ASTNSAATLHH715 pKa = 5.94VSATAAVPASTGTLVMNANGSYY737 pKa = 10.65VFTPASGFTGTLDD750 pKa = 3.36VAYY753 pKa = 8.0TVCDD757 pKa = 3.09NATPANCTEE766 pKa = 4.22ATLHH770 pKa = 5.74ITVDD774 pKa = 3.87PAPTATDD781 pKa = 3.78DD782 pKa = 4.73NIPGATNGVYY792 pKa = 7.25TTNILVNDD800 pKa = 4.36QIQPGAGISITRR812 pKa = 11.84QTGAGAGTAQGTVTFDD828 pKa = 3.75PLTGAMVYY836 pKa = 10.23VPSPLDD842 pKa = 3.7GNTVTVGYY850 pKa = 6.98TVCDD854 pKa = 3.58NNYY857 pKa = 10.21NPAQCADD864 pKa = 3.5ATVTIQVCDD873 pKa = 3.73PANPAMDD880 pKa = 4.65CDD882 pKa = 4.37GDD884 pKa = 4.18GVTNGQEE891 pKa = 4.35ILDD894 pKa = 4.18HH895 pKa = 6.89TDD897 pKa = 3.28PADD900 pKa = 3.88PCSLVAGSQTVAPKK914 pKa = 10.62GIWLTADD921 pKa = 3.46CDD923 pKa = 4.23GDD925 pKa = 4.07GVTNGQEE932 pKa = 4.43VIDD935 pKa = 4.27GTNPTDD941 pKa = 3.58PCDD944 pKa = 3.77YY945 pKa = 10.82NVASKK950 pKa = 9.26TLAPSAAWLALDD962 pKa = 4.59CDD964 pKa = 4.67GDD966 pKa = 4.06GTPNATDD973 pKa = 3.67PQPLNFCVGGTGAIPALGSATYY995 pKa = 11.04DD996 pKa = 3.22KK997 pKa = 10.88FFRR1000 pKa = 11.84NADD1003 pKa = 3.53CDD1005 pKa = 3.8GDD1007 pKa = 4.43GISNHH1012 pKa = 5.78MEE1014 pKa = 4.05TNTSGLALDD1023 pKa = 4.71FDD1025 pKa = 5.3GDD1027 pKa = 4.59GIPNYY1032 pKa = 10.67LDD1034 pKa = 3.98GDD1036 pKa = 4.12SDD1038 pKa = 4.37NDD1040 pKa = 4.68GIPDD1044 pKa = 3.42AVEE1047 pKa = 3.67QNRR1050 pKa = 11.84DD1051 pKa = 3.24SDD1053 pKa = 3.85GDD1055 pKa = 4.05GYY1057 pKa = 11.71ADD1059 pKa = 3.88YY1060 pKa = 11.33QDD1062 pKa = 5.3LDD1064 pKa = 3.89SDD1066 pKa = 4.06NDD1068 pKa = 4.66GISDD1072 pKa = 3.72KK1073 pKa = 11.29VEE1075 pKa = 4.42AGASPTMPVDD1085 pKa = 3.28SDD1087 pKa = 4.19GDD1089 pKa = 3.88GKK1091 pKa = 10.08PNYY1094 pKa = 10.03LDD1096 pKa = 5.17LDD1098 pKa = 3.91SDD1100 pKa = 3.87NDD1102 pKa = 4.66GILDD1106 pKa = 3.77SVEE1109 pKa = 4.68AIDD1112 pKa = 4.67VFAANRR1118 pKa = 11.84DD1119 pKa = 3.62DD1120 pKa = 5.2NYY1122 pKa = 11.27DD1123 pKa = 3.46GKK1125 pKa = 11.14VDD1127 pKa = 3.67KK1128 pKa = 11.36NGVFIDD1134 pKa = 3.81TNNNGWADD1142 pKa = 3.31ISEE1145 pKa = 4.32GDD1147 pKa = 3.99LPVDD1151 pKa = 3.83TDD1153 pKa = 3.55HH1154 pKa = 7.89DD1155 pKa = 5.17GIPDD1159 pKa = 3.88YY1160 pKa = 11.61LDD1162 pKa = 4.7LDD1164 pKa = 4.22SDD1166 pKa = 4.11NDD1168 pKa = 4.43CIPDD1172 pKa = 3.43AVEE1175 pKa = 4.18FTRR1178 pKa = 11.84DD1179 pKa = 2.79VDD1181 pKa = 3.53GDD1183 pKa = 3.47NRR1185 pKa = 11.84PNYY1188 pKa = 9.97RR1189 pKa = 11.84DD1190 pKa = 3.49TDD1192 pKa = 3.74SDD1194 pKa = 4.04GDD1196 pKa = 4.25GIPDD1200 pKa = 3.94NIEE1203 pKa = 4.3AGSCSHH1209 pKa = 7.65PIDD1212 pKa = 3.83TDD1214 pKa = 3.48GDD1216 pKa = 4.37GIPDD1220 pKa = 4.14FLDD1223 pKa = 3.49TDD1225 pKa = 4.07SDD1227 pKa = 3.99NDD1229 pKa = 4.73GIPDD1233 pKa = 4.73AIEE1236 pKa = 4.11AGKK1239 pKa = 10.33NPATPVDD1246 pKa = 3.75TDD1248 pKa = 3.42GDD1250 pKa = 4.5GIPDD1254 pKa = 3.78YY1255 pKa = 11.22RR1256 pKa = 11.84DD1257 pKa = 3.32LDD1259 pKa = 3.89SDD1261 pKa = 3.87NDD1263 pKa = 4.79GIPDD1267 pKa = 3.65ALEE1270 pKa = 4.09AGKK1273 pKa = 10.29DD1274 pKa = 3.42PKK1276 pKa = 11.32NPVDD1280 pKa = 3.62TDD1282 pKa = 3.65GDD1284 pKa = 4.08GTPDD1288 pKa = 3.9FQDD1291 pKa = 3.98LDD1293 pKa = 4.07SDD1295 pKa = 4.01NDD1297 pKa = 4.7GIPDD1301 pKa = 4.74AIEE1304 pKa = 4.07AGKK1307 pKa = 10.36DD1308 pKa = 3.16PKK1310 pKa = 11.32NPVDD1314 pKa = 3.7TDD1316 pKa = 3.5GDD1318 pKa = 4.39GIPDD1322 pKa = 4.01FRR1324 pKa = 11.84DD1325 pKa = 3.17LDD1327 pKa = 3.95SDD1329 pKa = 3.85NDD1331 pKa = 4.79GIPDD1335 pKa = 4.74AIEE1338 pKa = 4.07AGKK1341 pKa = 10.36DD1342 pKa = 3.16PKK1344 pKa = 11.32NPVDD1348 pKa = 3.7TDD1350 pKa = 3.5GDD1352 pKa = 4.39GIPDD1356 pKa = 4.01FRR1358 pKa = 11.84DD1359 pKa = 3.17LDD1361 pKa = 3.95SDD1363 pKa = 3.85NDD1365 pKa = 4.79GIPDD1369 pKa = 3.65ALEE1372 pKa = 4.09AGKK1375 pKa = 10.29DD1376 pKa = 3.42PKK1378 pKa = 11.32NPVDD1382 pKa = 3.7TDD1384 pKa = 3.5GDD1386 pKa = 4.39GIPDD1390 pKa = 3.72FRR1392 pKa = 11.84DD1393 pKa = 2.95VDD1395 pKa = 3.98SDD1397 pKa = 3.65NDD1399 pKa = 4.69GITDD1403 pKa = 3.6AFEE1406 pKa = 5.31AGANPKK1412 pKa = 10.5NPVDD1416 pKa = 3.74TDD1418 pKa = 3.5GDD1420 pKa = 4.39GIPDD1424 pKa = 4.01FRR1426 pKa = 11.84DD1427 pKa = 3.17LDD1429 pKa = 3.85SDD1431 pKa = 3.88NDD1433 pKa = 4.75GISDD1437 pKa = 4.2KK1438 pKa = 11.18IEE1440 pKa = 4.88GIVDD1444 pKa = 3.43TDD1446 pKa = 3.79KK1447 pKa = 11.72DD1448 pKa = 3.93GLPDD1452 pKa = 5.03FRR1454 pKa = 11.84DD1455 pKa = 3.54LDD1457 pKa = 4.28SDD1459 pKa = 3.77GDD1461 pKa = 3.99TYY1463 pKa = 10.97PDD1465 pKa = 3.5KK1466 pKa = 11.73VEE1468 pKa = 4.53GNVDD1472 pKa = 3.37TDD1474 pKa = 3.49KK1475 pKa = 11.74DD1476 pKa = 4.21GIPDD1480 pKa = 4.1FRR1482 pKa = 11.84DD1483 pKa = 3.2LDD1485 pKa = 3.78SDD1487 pKa = 4.17GDD1489 pKa = 4.1VIPDD1493 pKa = 3.75KK1494 pKa = 11.31LEE1496 pKa = 3.67NDD1498 pKa = 2.94IDD1500 pKa = 4.06YY1501 pKa = 11.37GGMPDD1506 pKa = 4.58CDD1508 pKa = 3.94GDD1510 pKa = 4.78GIDD1513 pKa = 5.04NRR1515 pKa = 11.84IDD1517 pKa = 3.77PDD1519 pKa = 3.78VCPTFATQGISPNNDD1534 pKa = 3.56GINDD1538 pKa = 3.51VLIIPGIKK1546 pKa = 9.57SYY1548 pKa = 11.17KK1549 pKa = 8.79NHH1551 pKa = 5.64LTIFNRR1557 pKa = 11.84WGNVVWEE1564 pKa = 4.13QEE1566 pKa = 4.04NYY1568 pKa = 8.04QNSWGGEE1575 pKa = 4.19TNQGDD1580 pKa = 4.31TVLSTDD1586 pKa = 4.13LKK1588 pKa = 11.13LPDD1591 pKa = 3.15GTYY1594 pKa = 10.4YY1595 pKa = 10.92YY1596 pKa = 11.06VIDD1599 pKa = 4.53FYY1601 pKa = 11.33GVKK1604 pKa = 9.98PNIGTYY1610 pKa = 10.5VYY1612 pKa = 10.65INRR1615 pKa = 11.84QAKK1618 pKa = 9.14
II1 pKa = 6.87ATVKK5 pKa = 10.41SGSSVTTNVLANDD18 pKa = 3.16VCEE21 pKa = 4.6NPNCTLSNPTIVTQPAHH38 pKa = 5.49GTVTVNPDD46 pKa = 2.68GTLTYY51 pKa = 10.04TPAAGFTGTDD61 pKa = 2.91VLTYY65 pKa = 7.88QVCDD69 pKa = 3.46NSVNPAVCKK78 pKa = 9.27TATVTYY84 pKa = 8.42TVQPATAAPTTTAADD99 pKa = 3.9DD100 pKa = 4.09ASTTNSNSPVSGNVLANDD118 pKa = 4.12GSTNPNAVLTVAPQNTTIPGKK139 pKa = 9.35GTIVMNADD147 pKa = 3.14GSFTFTPTGGFVGTVDD163 pKa = 3.99IPYY166 pKa = 8.67TVCDD170 pKa = 3.47NANPANCSTATLHH183 pKa = 6.73IVVEE187 pKa = 4.17PAPTVIAPDD196 pKa = 4.1FNNGLINKK204 pKa = 8.21PIAGSLATNDD214 pKa = 3.32VVAGGGTYY222 pKa = 7.65GTPVAATSNPTGATFTVNPDD242 pKa = 2.68GSYY245 pKa = 10.93SFTGTQPGVYY255 pKa = 8.66TYY257 pKa = 10.79NVPVCPAGQTTNCPLSPVTITVVDD281 pKa = 4.1PTAANVAPAVNPDD294 pKa = 2.88IATVKK299 pKa = 10.25SGSSVTTNVLANDD312 pKa = 3.16VCEE315 pKa = 4.6NPNCTLSNPTIVTQPAHH332 pKa = 5.49GTVTVNPDD340 pKa = 2.68GTLTYY345 pKa = 10.04TPAAGFTGTDD355 pKa = 2.91VLTYY359 pKa = 7.88QVCDD363 pKa = 3.46NSVNPAVCKK372 pKa = 9.92STTVTYY378 pKa = 9.08TVQPATAAPTTTAADD393 pKa = 3.79DD394 pKa = 3.93AASTKK399 pKa = 10.79GNVPATGNVLRR410 pKa = 11.84NDD412 pKa = 3.47ASTDD416 pKa = 3.45PNAVLTVTPQSTTIPGKK433 pKa = 9.33GTIVMNADD441 pKa = 2.99GSYY444 pKa = 9.39TFTPAPGFTGTVNIPYY460 pKa = 7.87TVCDD464 pKa = 3.41NANPANCSTATLHH477 pKa = 6.47VVVEE481 pKa = 4.32PPVDD485 pKa = 3.68LVGDD489 pKa = 4.16INATNINVPVNGNVSTNDD507 pKa = 3.4SAPGSVVYY515 pKa = 7.49GTPVASANNPSGATLTMNPDD535 pKa = 2.89GTYY538 pKa = 10.71SFTATTPGKK547 pKa = 10.31YY548 pKa = 10.27VYY550 pKa = 10.12DD551 pKa = 4.01VPNCLPGQTTGCPTTPLVITVKK573 pKa = 10.85DD574 pKa = 3.91PSATNNLPVVNPDD587 pKa = 2.8IATVRR592 pKa = 11.84AGTSVTTNVLANDD605 pKa = 3.07KK606 pKa = 9.13CTNQGCSLNPASVAITAQPAHH627 pKa = 6.01GTVTVNPDD635 pKa = 2.68GTLTYY640 pKa = 10.04TPAAGFTGTDD650 pKa = 2.87VLTYY654 pKa = 9.78KK655 pKa = 10.49VCDD658 pKa = 3.29NMTPANCQTATVTYY672 pKa = 8.71TVVPANSVLTTAADD686 pKa = 3.95DD687 pKa = 4.0YY688 pKa = 10.9VTTKK692 pKa = 9.69GTGIATGNVLGNDD705 pKa = 3.31ASTNSAATLHH715 pKa = 5.94VSATAAVPASTGTLVMNANGSYY737 pKa = 10.65VFTPASGFTGTLDD750 pKa = 3.36VAYY753 pKa = 8.0TVCDD757 pKa = 3.09NATPANCTEE766 pKa = 4.22ATLHH770 pKa = 5.74ITVDD774 pKa = 3.87PAPTATDD781 pKa = 3.78DD782 pKa = 4.73NIPGATNGVYY792 pKa = 7.25TTNILVNDD800 pKa = 4.36QIQPGAGISITRR812 pKa = 11.84QTGAGAGTAQGTVTFDD828 pKa = 3.75PLTGAMVYY836 pKa = 10.23VPSPLDD842 pKa = 3.7GNTVTVGYY850 pKa = 6.98TVCDD854 pKa = 3.58NNYY857 pKa = 10.21NPAQCADD864 pKa = 3.5ATVTIQVCDD873 pKa = 3.73PANPAMDD880 pKa = 4.65CDD882 pKa = 4.37GDD884 pKa = 4.18GVTNGQEE891 pKa = 4.35ILDD894 pKa = 4.18HH895 pKa = 6.89TDD897 pKa = 3.28PADD900 pKa = 3.88PCSLVAGSQTVAPKK914 pKa = 10.62GIWLTADD921 pKa = 3.46CDD923 pKa = 4.23GDD925 pKa = 4.07GVTNGQEE932 pKa = 4.43VIDD935 pKa = 4.27GTNPTDD941 pKa = 3.58PCDD944 pKa = 3.77YY945 pKa = 10.82NVASKK950 pKa = 9.26TLAPSAAWLALDD962 pKa = 4.59CDD964 pKa = 4.67GDD966 pKa = 4.06GTPNATDD973 pKa = 3.67PQPLNFCVGGTGAIPALGSATYY995 pKa = 11.04DD996 pKa = 3.22KK997 pKa = 10.88FFRR1000 pKa = 11.84NADD1003 pKa = 3.53CDD1005 pKa = 3.8GDD1007 pKa = 4.43GISNHH1012 pKa = 5.78MEE1014 pKa = 4.05TNTSGLALDD1023 pKa = 4.71FDD1025 pKa = 5.3GDD1027 pKa = 4.59GIPNYY1032 pKa = 10.67LDD1034 pKa = 3.98GDD1036 pKa = 4.12SDD1038 pKa = 4.37NDD1040 pKa = 4.68GIPDD1044 pKa = 3.42AVEE1047 pKa = 3.67QNRR1050 pKa = 11.84DD1051 pKa = 3.24SDD1053 pKa = 3.85GDD1055 pKa = 4.05GYY1057 pKa = 11.71ADD1059 pKa = 3.88YY1060 pKa = 11.33QDD1062 pKa = 5.3LDD1064 pKa = 3.89SDD1066 pKa = 4.06NDD1068 pKa = 4.66GISDD1072 pKa = 3.72KK1073 pKa = 11.29VEE1075 pKa = 4.42AGASPTMPVDD1085 pKa = 3.28SDD1087 pKa = 4.19GDD1089 pKa = 3.88GKK1091 pKa = 10.08PNYY1094 pKa = 10.03LDD1096 pKa = 5.17LDD1098 pKa = 3.91SDD1100 pKa = 3.87NDD1102 pKa = 4.66GILDD1106 pKa = 3.77SVEE1109 pKa = 4.68AIDD1112 pKa = 4.67VFAANRR1118 pKa = 11.84DD1119 pKa = 3.62DD1120 pKa = 5.2NYY1122 pKa = 11.27DD1123 pKa = 3.46GKK1125 pKa = 11.14VDD1127 pKa = 3.67KK1128 pKa = 11.36NGVFIDD1134 pKa = 3.81TNNNGWADD1142 pKa = 3.31ISEE1145 pKa = 4.32GDD1147 pKa = 3.99LPVDD1151 pKa = 3.83TDD1153 pKa = 3.55HH1154 pKa = 7.89DD1155 pKa = 5.17GIPDD1159 pKa = 3.88YY1160 pKa = 11.61LDD1162 pKa = 4.7LDD1164 pKa = 4.22SDD1166 pKa = 4.11NDD1168 pKa = 4.43CIPDD1172 pKa = 3.43AVEE1175 pKa = 4.18FTRR1178 pKa = 11.84DD1179 pKa = 2.79VDD1181 pKa = 3.53GDD1183 pKa = 3.47NRR1185 pKa = 11.84PNYY1188 pKa = 9.97RR1189 pKa = 11.84DD1190 pKa = 3.49TDD1192 pKa = 3.74SDD1194 pKa = 4.04GDD1196 pKa = 4.25GIPDD1200 pKa = 3.94NIEE1203 pKa = 4.3AGSCSHH1209 pKa = 7.65PIDD1212 pKa = 3.83TDD1214 pKa = 3.48GDD1216 pKa = 4.37GIPDD1220 pKa = 4.14FLDD1223 pKa = 3.49TDD1225 pKa = 4.07SDD1227 pKa = 3.99NDD1229 pKa = 4.73GIPDD1233 pKa = 4.73AIEE1236 pKa = 4.11AGKK1239 pKa = 10.33NPATPVDD1246 pKa = 3.75TDD1248 pKa = 3.42GDD1250 pKa = 4.5GIPDD1254 pKa = 3.78YY1255 pKa = 11.22RR1256 pKa = 11.84DD1257 pKa = 3.32LDD1259 pKa = 3.89SDD1261 pKa = 3.87NDD1263 pKa = 4.79GIPDD1267 pKa = 3.65ALEE1270 pKa = 4.09AGKK1273 pKa = 10.29DD1274 pKa = 3.42PKK1276 pKa = 11.32NPVDD1280 pKa = 3.62TDD1282 pKa = 3.65GDD1284 pKa = 4.08GTPDD1288 pKa = 3.9FQDD1291 pKa = 3.98LDD1293 pKa = 4.07SDD1295 pKa = 4.01NDD1297 pKa = 4.7GIPDD1301 pKa = 4.74AIEE1304 pKa = 4.07AGKK1307 pKa = 10.36DD1308 pKa = 3.16PKK1310 pKa = 11.32NPVDD1314 pKa = 3.7TDD1316 pKa = 3.5GDD1318 pKa = 4.39GIPDD1322 pKa = 4.01FRR1324 pKa = 11.84DD1325 pKa = 3.17LDD1327 pKa = 3.95SDD1329 pKa = 3.85NDD1331 pKa = 4.79GIPDD1335 pKa = 4.74AIEE1338 pKa = 4.07AGKK1341 pKa = 10.36DD1342 pKa = 3.16PKK1344 pKa = 11.32NPVDD1348 pKa = 3.7TDD1350 pKa = 3.5GDD1352 pKa = 4.39GIPDD1356 pKa = 4.01FRR1358 pKa = 11.84DD1359 pKa = 3.17LDD1361 pKa = 3.95SDD1363 pKa = 3.85NDD1365 pKa = 4.79GIPDD1369 pKa = 3.65ALEE1372 pKa = 4.09AGKK1375 pKa = 10.29DD1376 pKa = 3.42PKK1378 pKa = 11.32NPVDD1382 pKa = 3.7TDD1384 pKa = 3.5GDD1386 pKa = 4.39GIPDD1390 pKa = 3.72FRR1392 pKa = 11.84DD1393 pKa = 2.95VDD1395 pKa = 3.98SDD1397 pKa = 3.65NDD1399 pKa = 4.69GITDD1403 pKa = 3.6AFEE1406 pKa = 5.31AGANPKK1412 pKa = 10.5NPVDD1416 pKa = 3.74TDD1418 pKa = 3.5GDD1420 pKa = 4.39GIPDD1424 pKa = 4.01FRR1426 pKa = 11.84DD1427 pKa = 3.17LDD1429 pKa = 3.85SDD1431 pKa = 3.88NDD1433 pKa = 4.75GISDD1437 pKa = 4.2KK1438 pKa = 11.18IEE1440 pKa = 4.88GIVDD1444 pKa = 3.43TDD1446 pKa = 3.79KK1447 pKa = 11.72DD1448 pKa = 3.93GLPDD1452 pKa = 5.03FRR1454 pKa = 11.84DD1455 pKa = 3.54LDD1457 pKa = 4.28SDD1459 pKa = 3.77GDD1461 pKa = 3.99TYY1463 pKa = 10.97PDD1465 pKa = 3.5KK1466 pKa = 11.73VEE1468 pKa = 4.53GNVDD1472 pKa = 3.37TDD1474 pKa = 3.49KK1475 pKa = 11.74DD1476 pKa = 4.21GIPDD1480 pKa = 4.1FRR1482 pKa = 11.84DD1483 pKa = 3.2LDD1485 pKa = 3.78SDD1487 pKa = 4.17GDD1489 pKa = 4.1VIPDD1493 pKa = 3.75KK1494 pKa = 11.31LEE1496 pKa = 3.67NDD1498 pKa = 2.94IDD1500 pKa = 4.06YY1501 pKa = 11.37GGMPDD1506 pKa = 4.58CDD1508 pKa = 3.94GDD1510 pKa = 4.78GIDD1513 pKa = 5.04NRR1515 pKa = 11.84IDD1517 pKa = 3.77PDD1519 pKa = 3.78VCPTFATQGISPNNDD1534 pKa = 3.56GINDD1538 pKa = 3.51VLIIPGIKK1546 pKa = 9.57SYY1548 pKa = 11.17KK1549 pKa = 8.79NHH1551 pKa = 5.64LTIFNRR1557 pKa = 11.84WGNVVWEE1564 pKa = 4.13QEE1566 pKa = 4.04NYY1568 pKa = 8.04QNSWGGEE1575 pKa = 4.19TNQGDD1580 pKa = 4.31TVLSTDD1586 pKa = 4.13LKK1588 pKa = 11.13LPDD1591 pKa = 3.15GTYY1594 pKa = 10.4YY1595 pKa = 10.92YY1596 pKa = 11.06VIDD1599 pKa = 4.53FYY1601 pKa = 11.33GVKK1604 pKa = 9.98PNIGTYY1610 pKa = 10.5VYY1612 pKa = 10.65INRR1615 pKa = 11.84QAKK1618 pKa = 9.14
Molecular weight: 166.65 kDa
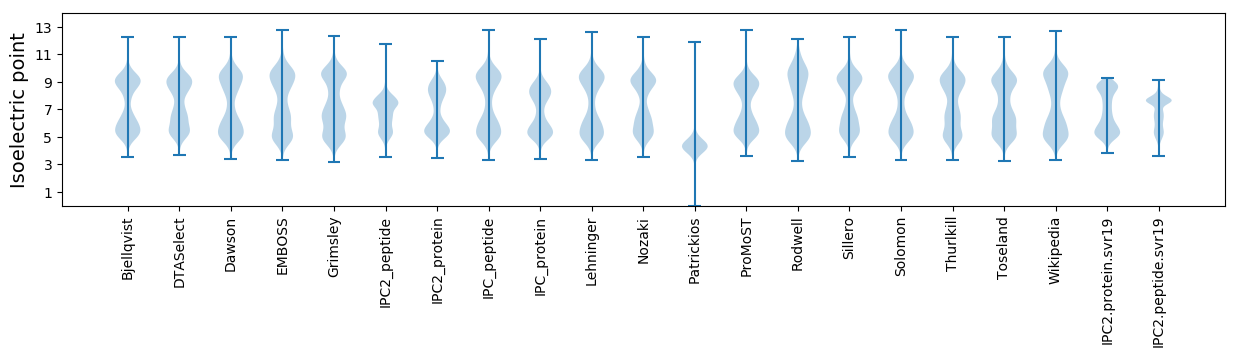
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A437PXL0|A0A437PXL0_9BACT FAD-binding protein OS=Cytophagaceae bacterium FSY-15 OX=1862386 GN=EOJ36_03135 PE=4 SV=1
MM1 pKa = 7.7KK2 pKa = 10.59NRR4 pKa = 11.84IFNLGFLVVFLFGITSCWSARR25 pKa = 11.84CPRR28 pKa = 11.84EE29 pKa = 3.74SCRR32 pKa = 11.84VQVEE36 pKa = 4.33HH37 pKa = 6.94RR38 pKa = 11.84HH39 pKa = 5.14GEE41 pKa = 3.96QYY43 pKa = 10.36FRR45 pKa = 11.84PRR47 pKa = 11.84AAFSWMYY54 pKa = 8.28TPRR57 pKa = 11.84YY58 pKa = 8.02KK59 pKa = 10.43HH60 pKa = 5.93IRR62 pKa = 11.84KK63 pKa = 9.46SPIAAGPNQPAVKK76 pKa = 10.45NPWYY80 pKa = 10.36KK81 pKa = 10.33FKK83 pKa = 11.0KK84 pKa = 8.64PSKK87 pKa = 8.99PASKK91 pKa = 10.19PRR93 pKa = 3.43
MM1 pKa = 7.7KK2 pKa = 10.59NRR4 pKa = 11.84IFNLGFLVVFLFGITSCWSARR25 pKa = 11.84CPRR28 pKa = 11.84EE29 pKa = 3.74SCRR32 pKa = 11.84VQVEE36 pKa = 4.33HH37 pKa = 6.94RR38 pKa = 11.84HH39 pKa = 5.14GEE41 pKa = 3.96QYY43 pKa = 10.36FRR45 pKa = 11.84PRR47 pKa = 11.84AAFSWMYY54 pKa = 8.28TPRR57 pKa = 11.84YY58 pKa = 8.02KK59 pKa = 10.43HH60 pKa = 5.93IRR62 pKa = 11.84KK63 pKa = 9.46SPIAAGPNQPAVKK76 pKa = 10.45NPWYY80 pKa = 10.36KK81 pKa = 10.33FKK83 pKa = 11.0KK84 pKa = 8.64PSKK87 pKa = 8.99PASKK91 pKa = 10.19PRR93 pKa = 3.43
Molecular weight: 10.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
835184 |
38 |
6376 |
360.0 |
40.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.843 ± 0.062 | 0.804 ± 0.017 |
4.967 ± 0.045 | 6.273 ± 0.074 |
5.367 ± 0.057 | 7.038 ± 0.058 |
1.796 ± 0.028 | 7.604 ± 0.057 |
7.636 ± 0.058 | 9.653 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.276 ± 0.033 | 5.745 ± 0.062 |
3.817 ± 0.033 | 3.912 ± 0.034 |
3.307 ± 0.035 | 6.721 ± 0.05 |
5.255 ± 0.101 | 6.056 ± 0.036 |
1.214 ± 0.018 | 3.713 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
