
Escherichia phage If1 (Bacteriophage If1)
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Infulavirus; Escherichia virus If1
Average proteome isoelectric point is 7.49
Get precalculated fractions of proteins
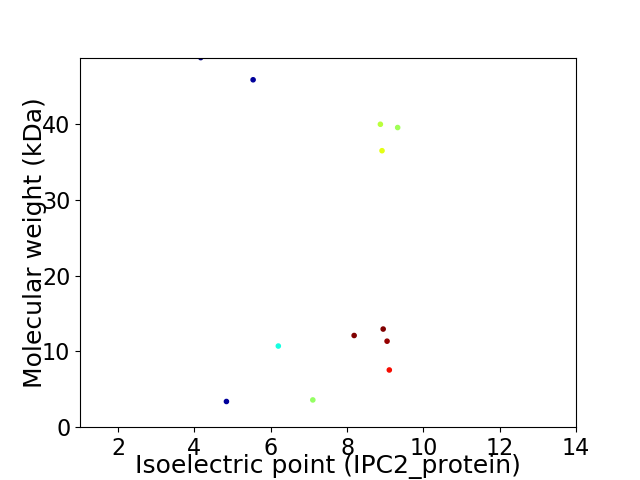
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
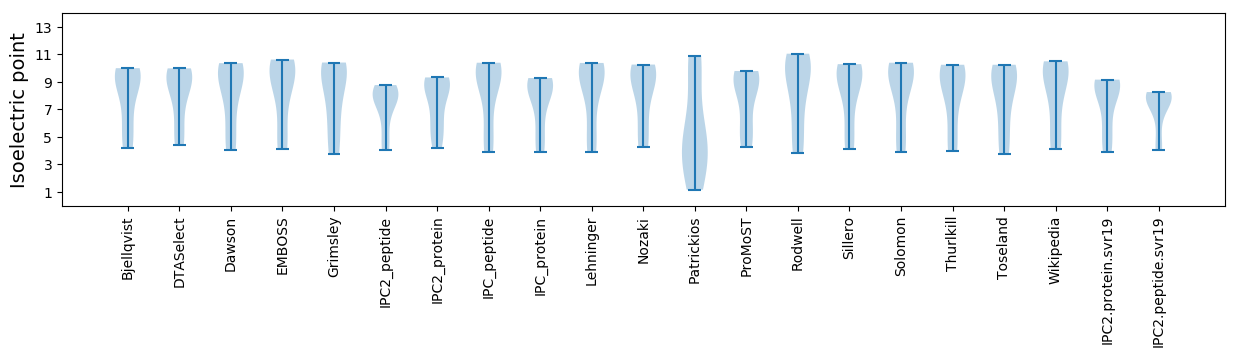
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|O80298|G6P_BPIF1 Head virion protein G6P OS=Escherichia phage If1 OX=10868 GN=VI PE=3 SV=1
MM1 pKa = 7.42KK2 pKa = 10.18KK3 pKa = 10.17IIIALFFAPFFTHH16 pKa = 6.46ATTDD20 pKa = 3.68AEE22 pKa = 4.63CLSKK26 pKa = 10.44PAFDD30 pKa = 3.9GTLSNVWKK38 pKa = 10.47EE39 pKa = 3.62GDD41 pKa = 3.17SRR43 pKa = 11.84YY44 pKa = 10.91ANFEE48 pKa = 3.69NCIYY52 pKa = 9.05EE53 pKa = 3.97LSGIGIGYY61 pKa = 10.46DD62 pKa = 3.38NDD64 pKa = 4.18TSCNGHH70 pKa = 5.18WTPVRR75 pKa = 11.84AADD78 pKa = 3.9GSGNGGDD85 pKa = 4.56DD86 pKa = 3.53NSSGGGSNGDD96 pKa = 3.44SGNNSTPDD104 pKa = 3.36TVTPGQTVNLPSDD117 pKa = 4.35LSTLSIPANVVKK129 pKa = 10.47SDD131 pKa = 4.09SIGSQFSLYY140 pKa = 10.11TNASCTMCSGYY151 pKa = 10.73YY152 pKa = 10.23LSNNADD158 pKa = 3.78SIAIANITEE167 pKa = 4.43TVKK170 pKa = 10.99ADD172 pKa = 3.48YY173 pKa = 9.49NQPDD177 pKa = 3.16MWFEE181 pKa = 4.04QTDD184 pKa = 3.28SDD186 pKa = 4.22GNHH189 pKa = 5.51VKK191 pKa = 10.27ILQNSYY197 pKa = 10.73KK198 pKa = 9.99AVSYY202 pKa = 10.6NVEE205 pKa = 4.35SKK207 pKa = 10.86QSDD210 pKa = 4.13VNNPTYY216 pKa = 10.65INYY219 pKa = 10.04SYY221 pKa = 11.1SVNVKK226 pKa = 9.34QVSYY230 pKa = 8.93DD231 pKa = 3.55TSNVCIMNWEE241 pKa = 4.28TFQNKK246 pKa = 8.99CDD248 pKa = 3.52ASRR251 pKa = 11.84AVLITDD257 pKa = 4.33TVTPSYY263 pKa = 10.82SRR265 pKa = 11.84NITIQSNINYY275 pKa = 9.43QGSNGSGGSGGSGGSGNDD293 pKa = 3.78GGGTGNNGNGTGDD306 pKa = 3.51FDD308 pKa = 4.26YY309 pKa = 11.8VKK311 pKa = 10.01MANANKK317 pKa = 10.17DD318 pKa = 3.36ALTEE322 pKa = 4.11SFDD325 pKa = 4.97LSALQADD332 pKa = 4.6TGASLDD338 pKa = 3.71GSVQGTLDD346 pKa = 3.73SLSGFSDD353 pKa = 3.8SIGGLVGNGSAISGEE368 pKa = 4.28FAGSSAAMNAIGEE381 pKa = 4.42GDD383 pKa = 3.95KK384 pKa = 11.47SPLLDD389 pKa = 3.62SLSFLKK395 pKa = 10.87DD396 pKa = 3.23GLFPALPEE404 pKa = 4.6FKK406 pKa = 10.21QCTPFVFAPGKK417 pKa = 8.92EE418 pKa = 3.82YY419 pKa = 11.02EE420 pKa = 4.78FIIEE424 pKa = 4.31CKK426 pKa = 10.2YY427 pKa = 10.49IDD429 pKa = 3.67MFKK432 pKa = 10.95GIFAFILYY440 pKa = 8.97FWTFVTVYY448 pKa = 10.71DD449 pKa = 4.09SFSGILRR456 pKa = 11.84KK457 pKa = 10.09GRR459 pKa = 11.84GG460 pKa = 3.19
MM1 pKa = 7.42KK2 pKa = 10.18KK3 pKa = 10.17IIIALFFAPFFTHH16 pKa = 6.46ATTDD20 pKa = 3.68AEE22 pKa = 4.63CLSKK26 pKa = 10.44PAFDD30 pKa = 3.9GTLSNVWKK38 pKa = 10.47EE39 pKa = 3.62GDD41 pKa = 3.17SRR43 pKa = 11.84YY44 pKa = 10.91ANFEE48 pKa = 3.69NCIYY52 pKa = 9.05EE53 pKa = 3.97LSGIGIGYY61 pKa = 10.46DD62 pKa = 3.38NDD64 pKa = 4.18TSCNGHH70 pKa = 5.18WTPVRR75 pKa = 11.84AADD78 pKa = 3.9GSGNGGDD85 pKa = 4.56DD86 pKa = 3.53NSSGGGSNGDD96 pKa = 3.44SGNNSTPDD104 pKa = 3.36TVTPGQTVNLPSDD117 pKa = 4.35LSTLSIPANVVKK129 pKa = 10.47SDD131 pKa = 4.09SIGSQFSLYY140 pKa = 10.11TNASCTMCSGYY151 pKa = 10.73YY152 pKa = 10.23LSNNADD158 pKa = 3.78SIAIANITEE167 pKa = 4.43TVKK170 pKa = 10.99ADD172 pKa = 3.48YY173 pKa = 9.49NQPDD177 pKa = 3.16MWFEE181 pKa = 4.04QTDD184 pKa = 3.28SDD186 pKa = 4.22GNHH189 pKa = 5.51VKK191 pKa = 10.27ILQNSYY197 pKa = 10.73KK198 pKa = 9.99AVSYY202 pKa = 10.6NVEE205 pKa = 4.35SKK207 pKa = 10.86QSDD210 pKa = 4.13VNNPTYY216 pKa = 10.65INYY219 pKa = 10.04SYY221 pKa = 11.1SVNVKK226 pKa = 9.34QVSYY230 pKa = 8.93DD231 pKa = 3.55TSNVCIMNWEE241 pKa = 4.28TFQNKK246 pKa = 8.99CDD248 pKa = 3.52ASRR251 pKa = 11.84AVLITDD257 pKa = 4.33TVTPSYY263 pKa = 10.82SRR265 pKa = 11.84NITIQSNINYY275 pKa = 9.43QGSNGSGGSGGSGGSGNDD293 pKa = 3.78GGGTGNNGNGTGDD306 pKa = 3.51FDD308 pKa = 4.26YY309 pKa = 11.8VKK311 pKa = 10.01MANANKK317 pKa = 10.17DD318 pKa = 3.36ALTEE322 pKa = 4.11SFDD325 pKa = 4.97LSALQADD332 pKa = 4.6TGASLDD338 pKa = 3.71GSVQGTLDD346 pKa = 3.73SLSGFSDD353 pKa = 3.8SIGGLVGNGSAISGEE368 pKa = 4.28FAGSSAAMNAIGEE381 pKa = 4.42GDD383 pKa = 3.95KK384 pKa = 11.47SPLLDD389 pKa = 3.62SLSFLKK395 pKa = 10.87DD396 pKa = 3.23GLFPALPEE404 pKa = 4.6FKK406 pKa = 10.21QCTPFVFAPGKK417 pKa = 8.92EE418 pKa = 3.82YY419 pKa = 11.02EE420 pKa = 4.78FIIEE424 pKa = 4.31CKK426 pKa = 10.2YY427 pKa = 10.49IDD429 pKa = 3.67MFKK432 pKa = 10.95GIFAFILYY440 pKa = 8.97FWTFVTVYY448 pKa = 10.71DD449 pKa = 4.09SFSGILRR456 pKa = 11.84KK457 pKa = 10.09GRR459 pKa = 11.84GG460 pKa = 3.19
Molecular weight: 48.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O80302|REP_BPIF1 Replication-associated protein G2P OS=Escherichia phage If1 OX=10868 GN=II PE=3 SV=1
MM1 pKa = 7.11WKK3 pKa = 10.27KK4 pKa = 8.54MPEE7 pKa = 4.07YY8 pKa = 10.44IFLQDD13 pKa = 4.06AYY15 pKa = 10.08TDD17 pKa = 3.7NLQQKK22 pKa = 10.34LKK24 pKa = 10.51DD25 pKa = 3.44LHH27 pKa = 5.86TAWKK31 pKa = 10.03RR32 pKa = 11.84CFDD35 pKa = 3.67KK36 pKa = 11.08KK37 pKa = 10.55LAAKK41 pKa = 10.4APVWKK46 pKa = 10.06RR47 pKa = 11.84KK48 pKa = 9.89NEE50 pKa = 3.75GRR52 pKa = 11.84DD53 pKa = 3.1SIRR56 pKa = 11.84FVNFEE61 pKa = 4.56KK62 pKa = 11.03YY63 pKa = 9.96CCLEE67 pKa = 3.61NRR69 pKa = 11.84RR70 pKa = 11.84VKK72 pKa = 10.73LPSGLGWVKK81 pKa = 10.01FRR83 pKa = 11.84QSQRR87 pKa = 11.84VNGKK91 pKa = 9.09IKK93 pKa = 10.68NATISQLAGQWYY105 pKa = 9.51ISFQVEE111 pKa = 3.95IEE113 pKa = 4.18TAEE116 pKa = 4.25PNHH119 pKa = 6.39TSTTIVGLDD128 pKa = 3.14AGVAKK133 pKa = 10.58LATLSDD139 pKa = 3.65GTVFEE144 pKa = 5.1PVNSFQKK151 pKa = 9.96NQKK154 pKa = 9.76KK155 pKa = 9.43LARR158 pKa = 11.84LQRR161 pKa = 11.84QLSRR165 pKa = 11.84KK166 pKa = 9.69VKK168 pKa = 10.59FSNNWQKK175 pKa = 10.65QKK177 pKa = 11.19RR178 pKa = 11.84KK179 pKa = 9.01IQCLHH184 pKa = 5.94SRR186 pKa = 11.84IANIRR191 pKa = 11.84RR192 pKa = 11.84DD193 pKa = 3.89YY194 pKa = 10.14LHH196 pKa = 7.11KK197 pKa = 10.62VTTTVSKK204 pKa = 8.75NHH206 pKa = 5.39AMIVIEE212 pKa = 4.52DD213 pKa = 3.94LKK215 pKa = 10.68VSNMSKK221 pKa = 10.13SAAGTVSQPGRR232 pKa = 11.84NVRR235 pKa = 11.84AKK237 pKa = 10.5SGLNRR242 pKa = 11.84SILDD246 pKa = 3.43QGWYY250 pKa = 9.21EE251 pKa = 3.93MRR253 pKa = 11.84RR254 pKa = 11.84QLEE257 pKa = 4.23YY258 pKa = 10.35KK259 pKa = 9.57QLWRR263 pKa = 11.84GGQVLAVPPAYY274 pKa = 9.4TSQRR278 pKa = 11.84CACCGHH284 pKa = 5.45TAKK287 pKa = 10.5EE288 pKa = 4.1NRR290 pKa = 11.84LSQSQFRR297 pKa = 11.84CQVCGYY303 pKa = 6.91TANADD308 pKa = 3.46VNGARR313 pKa = 11.84NILAAGHH320 pKa = 6.74AVLACGGTMQSDD332 pKa = 3.59RR333 pKa = 11.84PLKK336 pKa = 10.19QEE338 pKa = 3.99PDD340 pKa = 3.51AEE342 pKa = 4.31QSAVTHH348 pKa = 6.62
MM1 pKa = 7.11WKK3 pKa = 10.27KK4 pKa = 8.54MPEE7 pKa = 4.07YY8 pKa = 10.44IFLQDD13 pKa = 4.06AYY15 pKa = 10.08TDD17 pKa = 3.7NLQQKK22 pKa = 10.34LKK24 pKa = 10.51DD25 pKa = 3.44LHH27 pKa = 5.86TAWKK31 pKa = 10.03RR32 pKa = 11.84CFDD35 pKa = 3.67KK36 pKa = 11.08KK37 pKa = 10.55LAAKK41 pKa = 10.4APVWKK46 pKa = 10.06RR47 pKa = 11.84KK48 pKa = 9.89NEE50 pKa = 3.75GRR52 pKa = 11.84DD53 pKa = 3.1SIRR56 pKa = 11.84FVNFEE61 pKa = 4.56KK62 pKa = 11.03YY63 pKa = 9.96CCLEE67 pKa = 3.61NRR69 pKa = 11.84RR70 pKa = 11.84VKK72 pKa = 10.73LPSGLGWVKK81 pKa = 10.01FRR83 pKa = 11.84QSQRR87 pKa = 11.84VNGKK91 pKa = 9.09IKK93 pKa = 10.68NATISQLAGQWYY105 pKa = 9.51ISFQVEE111 pKa = 3.95IEE113 pKa = 4.18TAEE116 pKa = 4.25PNHH119 pKa = 6.39TSTTIVGLDD128 pKa = 3.14AGVAKK133 pKa = 10.58LATLSDD139 pKa = 3.65GTVFEE144 pKa = 5.1PVNSFQKK151 pKa = 9.96NQKK154 pKa = 9.76KK155 pKa = 9.43LARR158 pKa = 11.84LQRR161 pKa = 11.84QLSRR165 pKa = 11.84KK166 pKa = 9.69VKK168 pKa = 10.59FSNNWQKK175 pKa = 10.65QKK177 pKa = 11.19RR178 pKa = 11.84KK179 pKa = 9.01IQCLHH184 pKa = 5.94SRR186 pKa = 11.84IANIRR191 pKa = 11.84RR192 pKa = 11.84DD193 pKa = 3.89YY194 pKa = 10.14LHH196 pKa = 7.11KK197 pKa = 10.62VTTTVSKK204 pKa = 8.75NHH206 pKa = 5.39AMIVIEE212 pKa = 4.52DD213 pKa = 3.94LKK215 pKa = 10.68VSNMSKK221 pKa = 10.13SAAGTVSQPGRR232 pKa = 11.84NVRR235 pKa = 11.84AKK237 pKa = 10.5SGLNRR242 pKa = 11.84SILDD246 pKa = 3.43QGWYY250 pKa = 9.21EE251 pKa = 3.93MRR253 pKa = 11.84RR254 pKa = 11.84QLEE257 pKa = 4.23YY258 pKa = 10.35KK259 pKa = 9.57QLWRR263 pKa = 11.84GGQVLAVPPAYY274 pKa = 9.4TSQRR278 pKa = 11.84CACCGHH284 pKa = 5.45TAKK287 pKa = 10.5EE288 pKa = 4.1NRR290 pKa = 11.84LSQSQFRR297 pKa = 11.84CQVCGYY303 pKa = 6.91TANADD308 pKa = 3.46VNGARR313 pKa = 11.84NILAAGHH320 pKa = 6.74AVLACGGTMQSDD332 pKa = 3.59RR333 pKa = 11.84PLKK336 pKa = 10.19QEE338 pKa = 3.99PDD340 pKa = 3.51AEE342 pKa = 4.31QSAVTHH348 pKa = 6.62
Molecular weight: 39.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2471 |
32 |
460 |
205.9 |
22.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.365 ± 0.703 | 1.7 ± 0.341 |
5.301 ± 0.556 | 3.642 ± 0.336 |
4.816 ± 0.391 | 7.285 ± 0.845 |
1.538 ± 0.406 | 6.394 ± 0.469 |
6.677 ± 0.731 | 8.175 ± 0.729 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.781 ± 0.146 | 4.978 ± 0.939 |
3.885 ± 0.569 | 3.561 ± 0.729 |
4.573 ± 0.853 | 9.51 ± 0.927 |
6.273 ± 0.634 | 7.689 ± 0.663 |
1.255 ± 0.257 | 3.602 ± 0.454 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
