
Pteromalus puparum negative-strand RNA virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Artoviridae; Peropuvirus; Pteromalus puparum peropuvirus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
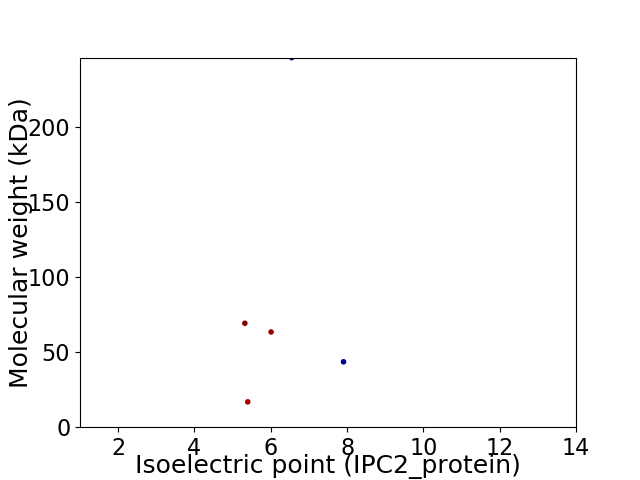
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L5BWP3|A0A1L5BWP3_9MONO Uncharacterized protein OS=Pteromalus puparum negative-strand RNA virus 1 OX=1926633 PE=4 SV=1
MM1 pKa = 7.3PVVVPSNSAMAFTYY15 pKa = 11.11ADD17 pKa = 4.75LNPLLQAAGFPSIPDD32 pKa = 3.33NTTSWRR38 pKa = 11.84FTSYY42 pKa = 11.07KK43 pKa = 10.67DD44 pKa = 3.8LISFRR49 pKa = 11.84ARR51 pKa = 11.84ILPTTIGMMIDD62 pKa = 3.23QATPPRR68 pKa = 11.84VDD70 pKa = 2.89GAQFEE75 pKa = 5.02QNSSTAPPNSYY86 pKa = 10.5LYY88 pKa = 10.69FIATVLKK95 pKa = 9.66RR96 pKa = 11.84TDD98 pKa = 2.84VDD100 pKa = 3.64YY101 pKa = 11.28TGILVAVGLPQFCASVKK118 pKa = 9.5RR119 pKa = 11.84TLLSLQPEE127 pKa = 4.46LEE129 pKa = 3.89ILANEE134 pKa = 4.47DD135 pKa = 3.32EE136 pKa = 4.9LNRR139 pKa = 11.84FPPLTGIRR147 pKa = 11.84QFTSLGATEE156 pKa = 4.55LTCFLASEE164 pKa = 4.2DD165 pKa = 3.82HH166 pKa = 6.62LFQILEE172 pKa = 4.3DD173 pKa = 3.52QTISQEE179 pKa = 4.28SRR181 pKa = 11.84VCTALVAMFPYY192 pKa = 10.34ILPEE196 pKa = 3.95LQTRR200 pKa = 11.84VHH202 pKa = 6.27VVHH205 pKa = 7.17TYY207 pKa = 10.21QGGAQVRR214 pKa = 11.84PEE216 pKa = 3.8ALFGGGAEE224 pKa = 3.94QAVEE228 pKa = 3.75NARR231 pKa = 11.84RR232 pKa = 11.84LTLLAIYY239 pKa = 10.02LLAPAYY245 pKa = 8.36TRR247 pKa = 11.84QDD249 pKa = 3.31KK250 pKa = 10.63LHH252 pKa = 5.78EE253 pKa = 4.41QIARR257 pKa = 11.84RR258 pKa = 11.84TQAAAVQEE266 pKa = 4.18QLEE269 pKa = 4.77HH270 pKa = 6.47YY271 pKa = 8.54DD272 pKa = 3.24ASRR275 pKa = 11.84TRR277 pKa = 11.84TLLDD281 pKa = 3.18SLQWKK286 pKa = 8.52IGSMMPIVDD295 pKa = 4.74DD296 pKa = 3.77VLAMVICRR304 pKa = 11.84TTRR307 pKa = 11.84GTTDD311 pKa = 3.21PNAPLTEE318 pKa = 5.11LISPGEE324 pKa = 3.88LNRR327 pKa = 11.84NQEE330 pKa = 3.99IGRR333 pKa = 11.84VASSLLEE340 pKa = 4.01QARR343 pKa = 11.84LVYY346 pKa = 10.05TNYY349 pKa = 10.14HH350 pKa = 5.08STSIHH355 pKa = 6.57FILPVFDD362 pKa = 4.89PLVTAIGADD371 pKa = 2.81HH372 pKa = 6.27LTFRR376 pKa = 11.84EE377 pKa = 4.12EE378 pKa = 3.34ITRR381 pKa = 11.84FRR383 pKa = 11.84GIRR386 pKa = 11.84TVVSSAPFMGLCAQLPAQYY405 pKa = 10.03HH406 pKa = 5.94IKK408 pKa = 10.3NCARR412 pKa = 11.84LAYY415 pKa = 10.38LGLLYY420 pKa = 10.19HH421 pKa = 6.98DD422 pKa = 5.14RR423 pKa = 11.84SLTTPEE429 pKa = 3.79EE430 pKa = 3.91KK431 pKa = 10.68ASFKK435 pKa = 10.24EE436 pKa = 4.12YY437 pKa = 10.22KK438 pKa = 9.57ISGVRR443 pKa = 11.84EE444 pKa = 4.38HH445 pKa = 7.04IASQADD451 pKa = 3.89RR452 pKa = 11.84NIVEE456 pKa = 4.33NLVDD460 pKa = 3.92ILPSQTVSAIASLVQHH476 pKa = 6.77ISLEE480 pKa = 4.19KK481 pKa = 10.68ASILMDD487 pKa = 3.63SKK489 pKa = 11.42SPEE492 pKa = 3.89EE493 pKa = 3.76QLEE496 pKa = 4.15VLNLLRR502 pKa = 11.84GEE504 pKa = 4.47DD505 pKa = 4.28APGAWAMDD513 pKa = 3.29QMNRR517 pKa = 11.84EE518 pKa = 3.85NATYY522 pKa = 9.24TRR524 pKa = 11.84QLIEE528 pKa = 4.55SGVKK532 pKa = 9.68LLKK535 pKa = 10.46AALEE539 pKa = 4.08EE540 pKa = 4.23QYY542 pKa = 11.14EE543 pKa = 4.18LRR545 pKa = 11.84RR546 pKa = 11.84DD547 pKa = 3.56AAEE550 pKa = 4.02TLPTPDD556 pKa = 2.82AVRR559 pKa = 11.84TRR561 pKa = 11.84RR562 pKa = 11.84AALIEE567 pKa = 3.83WRR569 pKa = 11.84KK570 pKa = 10.48GITNMFSNLTDD581 pKa = 3.09WTEE584 pKa = 4.32ALPSAPRR591 pKa = 11.84SGMEE595 pKa = 4.77AIRR598 pKa = 11.84TTTLARR604 pKa = 11.84LTEE607 pKa = 3.97IMTAIRR613 pKa = 11.84TDD615 pKa = 3.31DD616 pKa = 3.86AMEE619 pKa = 3.79QQ620 pKa = 3.35
MM1 pKa = 7.3PVVVPSNSAMAFTYY15 pKa = 11.11ADD17 pKa = 4.75LNPLLQAAGFPSIPDD32 pKa = 3.33NTTSWRR38 pKa = 11.84FTSYY42 pKa = 11.07KK43 pKa = 10.67DD44 pKa = 3.8LISFRR49 pKa = 11.84ARR51 pKa = 11.84ILPTTIGMMIDD62 pKa = 3.23QATPPRR68 pKa = 11.84VDD70 pKa = 2.89GAQFEE75 pKa = 5.02QNSSTAPPNSYY86 pKa = 10.5LYY88 pKa = 10.69FIATVLKK95 pKa = 9.66RR96 pKa = 11.84TDD98 pKa = 2.84VDD100 pKa = 3.64YY101 pKa = 11.28TGILVAVGLPQFCASVKK118 pKa = 9.5RR119 pKa = 11.84TLLSLQPEE127 pKa = 4.46LEE129 pKa = 3.89ILANEE134 pKa = 4.47DD135 pKa = 3.32EE136 pKa = 4.9LNRR139 pKa = 11.84FPPLTGIRR147 pKa = 11.84QFTSLGATEE156 pKa = 4.55LTCFLASEE164 pKa = 4.2DD165 pKa = 3.82HH166 pKa = 6.62LFQILEE172 pKa = 4.3DD173 pKa = 3.52QTISQEE179 pKa = 4.28SRR181 pKa = 11.84VCTALVAMFPYY192 pKa = 10.34ILPEE196 pKa = 3.95LQTRR200 pKa = 11.84VHH202 pKa = 6.27VVHH205 pKa = 7.17TYY207 pKa = 10.21QGGAQVRR214 pKa = 11.84PEE216 pKa = 3.8ALFGGGAEE224 pKa = 3.94QAVEE228 pKa = 3.75NARR231 pKa = 11.84RR232 pKa = 11.84LTLLAIYY239 pKa = 10.02LLAPAYY245 pKa = 8.36TRR247 pKa = 11.84QDD249 pKa = 3.31KK250 pKa = 10.63LHH252 pKa = 5.78EE253 pKa = 4.41QIARR257 pKa = 11.84RR258 pKa = 11.84TQAAAVQEE266 pKa = 4.18QLEE269 pKa = 4.77HH270 pKa = 6.47YY271 pKa = 8.54DD272 pKa = 3.24ASRR275 pKa = 11.84TRR277 pKa = 11.84TLLDD281 pKa = 3.18SLQWKK286 pKa = 8.52IGSMMPIVDD295 pKa = 4.74DD296 pKa = 3.77VLAMVICRR304 pKa = 11.84TTRR307 pKa = 11.84GTTDD311 pKa = 3.21PNAPLTEE318 pKa = 5.11LISPGEE324 pKa = 3.88LNRR327 pKa = 11.84NQEE330 pKa = 3.99IGRR333 pKa = 11.84VASSLLEE340 pKa = 4.01QARR343 pKa = 11.84LVYY346 pKa = 10.05TNYY349 pKa = 10.14HH350 pKa = 5.08STSIHH355 pKa = 6.57FILPVFDD362 pKa = 4.89PLVTAIGADD371 pKa = 2.81HH372 pKa = 6.27LTFRR376 pKa = 11.84EE377 pKa = 4.12EE378 pKa = 3.34ITRR381 pKa = 11.84FRR383 pKa = 11.84GIRR386 pKa = 11.84TVVSSAPFMGLCAQLPAQYY405 pKa = 10.03HH406 pKa = 5.94IKK408 pKa = 10.3NCARR412 pKa = 11.84LAYY415 pKa = 10.38LGLLYY420 pKa = 10.19HH421 pKa = 6.98DD422 pKa = 5.14RR423 pKa = 11.84SLTTPEE429 pKa = 3.79EE430 pKa = 3.91KK431 pKa = 10.68ASFKK435 pKa = 10.24EE436 pKa = 4.12YY437 pKa = 10.22KK438 pKa = 9.57ISGVRR443 pKa = 11.84EE444 pKa = 4.38HH445 pKa = 7.04IASQADD451 pKa = 3.89RR452 pKa = 11.84NIVEE456 pKa = 4.33NLVDD460 pKa = 3.92ILPSQTVSAIASLVQHH476 pKa = 6.77ISLEE480 pKa = 4.19KK481 pKa = 10.68ASILMDD487 pKa = 3.63SKK489 pKa = 11.42SPEE492 pKa = 3.89EE493 pKa = 3.76QLEE496 pKa = 4.15VLNLLRR502 pKa = 11.84GEE504 pKa = 4.47DD505 pKa = 4.28APGAWAMDD513 pKa = 3.29QMNRR517 pKa = 11.84EE518 pKa = 3.85NATYY522 pKa = 9.24TRR524 pKa = 11.84QLIEE528 pKa = 4.55SGVKK532 pKa = 9.68LLKK535 pKa = 10.46AALEE539 pKa = 4.08EE540 pKa = 4.23QYY542 pKa = 11.14EE543 pKa = 4.18LRR545 pKa = 11.84RR546 pKa = 11.84DD547 pKa = 3.56AAEE550 pKa = 4.02TLPTPDD556 pKa = 2.82AVRR559 pKa = 11.84TRR561 pKa = 11.84RR562 pKa = 11.84AALIEE567 pKa = 3.83WRR569 pKa = 11.84KK570 pKa = 10.48GITNMFSNLTDD581 pKa = 3.09WTEE584 pKa = 4.32ALPSAPRR591 pKa = 11.84SGMEE595 pKa = 4.77AIRR598 pKa = 11.84TTTLARR604 pKa = 11.84LTEE607 pKa = 3.97IMTAIRR613 pKa = 11.84TDD615 pKa = 3.31DD616 pKa = 3.86AMEE619 pKa = 3.79QQ620 pKa = 3.35
Molecular weight: 69.13 kDa
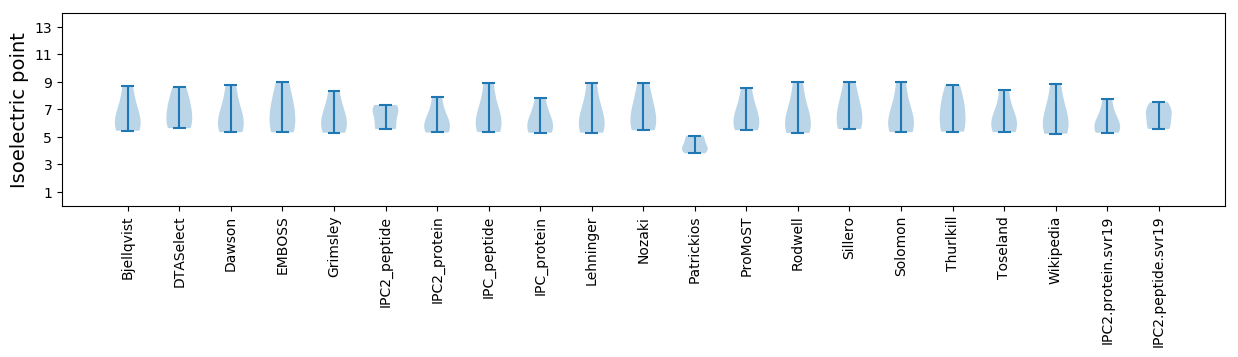
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L5BWP9|A0A1L5BWP9_9MONO GDP polyribonucleotidyltransferase OS=Pteromalus puparum negative-strand RNA virus 1 OX=1926633 PE=4 SV=1
MM1 pKa = 7.46SGKK4 pKa = 10.02KK5 pKa = 9.39DD6 pKa = 3.27AKK8 pKa = 9.41TPRR11 pKa = 11.84SATKK15 pKa = 8.89GTLSKK20 pKa = 11.17AEE22 pKa = 4.29EE23 pKa = 4.27LLQQLAAYY31 pKa = 9.53DD32 pKa = 4.13PLTNNDD38 pKa = 4.15PLSLGKK44 pKa = 10.02KK45 pKa = 8.8RR46 pKa = 11.84PKK48 pKa = 9.91PPKK51 pKa = 10.27CPDD54 pKa = 2.89IQEE57 pKa = 4.89PSDD60 pKa = 3.52SPPYY64 pKa = 9.59KK65 pKa = 10.31RR66 pKa = 11.84HH67 pKa = 5.71DD68 pKa = 3.7TRR70 pKa = 11.84GDD72 pKa = 3.53EE73 pKa = 4.33PDD75 pKa = 3.33EE76 pKa = 4.37TRR78 pKa = 11.84DD79 pKa = 3.92PDD81 pKa = 4.15PSLHH85 pKa = 6.16PCYY88 pKa = 10.43YY89 pKa = 9.93PLGNSSSTASIVEE102 pKa = 4.43VIEE105 pKa = 5.57SDD107 pKa = 4.04SQDD110 pKa = 3.58LNPPEE115 pKa = 4.19EE116 pKa = 4.51PKK118 pKa = 10.77DD119 pKa = 3.83RR120 pKa = 11.84PSTRR124 pKa = 11.84GEE126 pKa = 3.94TSRR129 pKa = 11.84TATRR133 pKa = 11.84EE134 pKa = 3.7QRR136 pKa = 11.84EE137 pKa = 4.08VLSIRR142 pKa = 11.84KK143 pKa = 8.65LADD146 pKa = 3.33TQEE149 pKa = 4.53VISSDD154 pKa = 3.45EE155 pKa = 4.1EE156 pKa = 4.25EE157 pKa = 4.31EE158 pKa = 4.31GRR160 pKa = 11.84EE161 pKa = 3.98SGAVPKK167 pKa = 10.66GFYY170 pKa = 9.31TVPPPVLQYY179 pKa = 9.6ITGEE183 pKa = 4.18SSKK186 pKa = 10.96KK187 pKa = 9.71PGGFQTEE194 pKa = 3.77MDD196 pKa = 3.08RR197 pKa = 11.84WFEE200 pKa = 4.17DD201 pKa = 3.03EE202 pKa = 3.98WGKK205 pKa = 10.34LVNPPPGLLSTHH217 pKa = 6.14YY218 pKa = 9.84LWCLPPSTTNTLAGEE233 pKa = 4.21KK234 pKa = 10.42FKK236 pKa = 11.32VASNNLAISLGRR248 pKa = 11.84SKK250 pKa = 10.23IWGKK254 pKa = 10.51KK255 pKa = 8.98KK256 pKa = 10.18SSKK259 pKa = 8.92GTAGVLVLPFPGVQSNTAEE278 pKa = 4.21KK279 pKa = 10.55AAGDD283 pKa = 4.18RR284 pKa = 11.84LTKK287 pKa = 10.36AAEE290 pKa = 4.09ALQQLAEE297 pKa = 4.4DD298 pKa = 4.77FKK300 pKa = 11.54SAIAILQADD309 pKa = 4.51INRR312 pKa = 11.84QAAASRR318 pKa = 11.84GFITDD323 pKa = 3.42SSQSIRR329 pKa = 11.84ANTLSLIRR337 pKa = 11.84SMEE340 pKa = 3.95EE341 pKa = 3.31TAKK344 pKa = 10.46SVKK347 pKa = 9.33GTTTPVQGMGEE358 pKa = 4.11SRR360 pKa = 11.84RR361 pKa = 11.84DD362 pKa = 3.39SRR364 pKa = 11.84TTTPSLPGPGAGQSSRR380 pKa = 11.84PSTGGLTIDD389 pKa = 3.28PSKK392 pKa = 10.64FKK394 pKa = 10.73RR395 pKa = 11.84IVPQGKK401 pKa = 9.04KK402 pKa = 9.82
MM1 pKa = 7.46SGKK4 pKa = 10.02KK5 pKa = 9.39DD6 pKa = 3.27AKK8 pKa = 9.41TPRR11 pKa = 11.84SATKK15 pKa = 8.89GTLSKK20 pKa = 11.17AEE22 pKa = 4.29EE23 pKa = 4.27LLQQLAAYY31 pKa = 9.53DD32 pKa = 4.13PLTNNDD38 pKa = 4.15PLSLGKK44 pKa = 10.02KK45 pKa = 8.8RR46 pKa = 11.84PKK48 pKa = 9.91PPKK51 pKa = 10.27CPDD54 pKa = 2.89IQEE57 pKa = 4.89PSDD60 pKa = 3.52SPPYY64 pKa = 9.59KK65 pKa = 10.31RR66 pKa = 11.84HH67 pKa = 5.71DD68 pKa = 3.7TRR70 pKa = 11.84GDD72 pKa = 3.53EE73 pKa = 4.33PDD75 pKa = 3.33EE76 pKa = 4.37TRR78 pKa = 11.84DD79 pKa = 3.92PDD81 pKa = 4.15PSLHH85 pKa = 6.16PCYY88 pKa = 10.43YY89 pKa = 9.93PLGNSSSTASIVEE102 pKa = 4.43VIEE105 pKa = 5.57SDD107 pKa = 4.04SQDD110 pKa = 3.58LNPPEE115 pKa = 4.19EE116 pKa = 4.51PKK118 pKa = 10.77DD119 pKa = 3.83RR120 pKa = 11.84PSTRR124 pKa = 11.84GEE126 pKa = 3.94TSRR129 pKa = 11.84TATRR133 pKa = 11.84EE134 pKa = 3.7QRR136 pKa = 11.84EE137 pKa = 4.08VLSIRR142 pKa = 11.84KK143 pKa = 8.65LADD146 pKa = 3.33TQEE149 pKa = 4.53VISSDD154 pKa = 3.45EE155 pKa = 4.1EE156 pKa = 4.25EE157 pKa = 4.31EE158 pKa = 4.31GRR160 pKa = 11.84EE161 pKa = 3.98SGAVPKK167 pKa = 10.66GFYY170 pKa = 9.31TVPPPVLQYY179 pKa = 9.6ITGEE183 pKa = 4.18SSKK186 pKa = 10.96KK187 pKa = 9.71PGGFQTEE194 pKa = 3.77MDD196 pKa = 3.08RR197 pKa = 11.84WFEE200 pKa = 4.17DD201 pKa = 3.03EE202 pKa = 3.98WGKK205 pKa = 10.34LVNPPPGLLSTHH217 pKa = 6.14YY218 pKa = 9.84LWCLPPSTTNTLAGEE233 pKa = 4.21KK234 pKa = 10.42FKK236 pKa = 11.32VASNNLAISLGRR248 pKa = 11.84SKK250 pKa = 10.23IWGKK254 pKa = 10.51KK255 pKa = 8.98KK256 pKa = 10.18SSKK259 pKa = 8.92GTAGVLVLPFPGVQSNTAEE278 pKa = 4.21KK279 pKa = 10.55AAGDD283 pKa = 4.18RR284 pKa = 11.84LTKK287 pKa = 10.36AAEE290 pKa = 4.09ALQQLAEE297 pKa = 4.4DD298 pKa = 4.77FKK300 pKa = 11.54SAIAILQADD309 pKa = 4.51INRR312 pKa = 11.84QAAASRR318 pKa = 11.84GFITDD323 pKa = 3.42SSQSIRR329 pKa = 11.84ANTLSLIRR337 pKa = 11.84SMEE340 pKa = 3.95EE341 pKa = 3.31TAKK344 pKa = 10.46SVKK347 pKa = 9.33GTTTPVQGMGEE358 pKa = 4.11SRR360 pKa = 11.84RR361 pKa = 11.84DD362 pKa = 3.39SRR364 pKa = 11.84TTTPSLPGPGAGQSSRR380 pKa = 11.84PSTGGLTIDD389 pKa = 3.28PSKK392 pKa = 10.64FKK394 pKa = 10.73RR395 pKa = 11.84IVPQGKK401 pKa = 9.04KK402 pKa = 9.82
Molecular weight: 43.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3913 |
151 |
2166 |
782.6 |
87.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.645 ± 0.951 | 1.508 ± 0.277 |
4.983 ± 0.309 | 5.929 ± 0.613 |
3.271 ± 0.706 | 5.929 ± 0.568 |
2.326 ± 0.505 | 6.21 ± 0.444 |
4.779 ± 0.694 | 11.756 ± 0.999 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.249 ± 0.237 | 3.603 ± 0.25 |
5.443 ± 0.79 | 4.651 ± 0.132 |
5.597 ± 0.489 | 7.335 ± 0.865 |
7.105 ± 0.985 | 5.955 ± 0.573 |
1.482 ± 0.353 | 3.246 ± 0.535 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
