
Aquimarina algiphila
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aquimarina
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
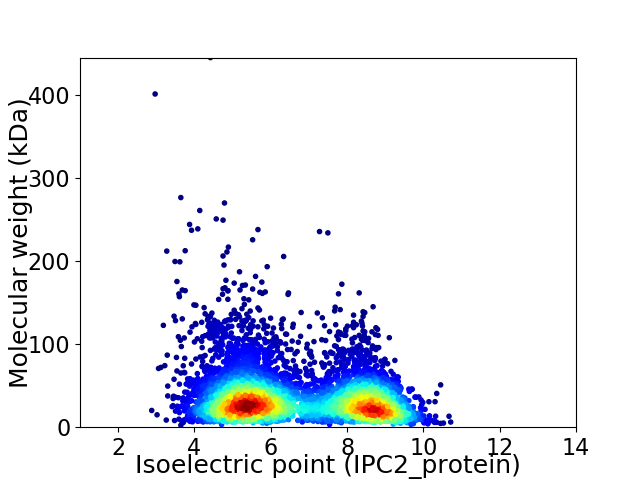
Virtual 2D-PAGE plot for 6064 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A554VIL8|A0A554VIL8_9FLAO Uncharacterized protein OS=Aquimarina algiphila OX=2047982 GN=FOF46_15305 PE=4 SV=1
MM1 pKa = 7.3NKK3 pKa = 8.26KK4 pKa = 8.54TYY6 pKa = 10.25LIVVILLTISKK17 pKa = 10.41GLFAQTMTLTSNAPSNTICRR37 pKa = 11.84GDD39 pKa = 3.85IIQVDD44 pKa = 4.21VNITPPNGASGYY56 pKa = 9.43NWTNVTTGQFIKK68 pKa = 10.62SVNGNAGTFHH78 pKa = 7.52LFTGVDD84 pKa = 3.27RR85 pKa = 11.84LLTNSTIRR93 pKa = 11.84VEE95 pKa = 4.4AFGGTGGNLSQTIDD109 pKa = 3.85IIVQQPANPGTEE121 pKa = 4.24GNLLLCNKK129 pKa = 9.35TGDD132 pKa = 3.39IDD134 pKa = 6.3LFTLLQGNPDD144 pKa = 4.2RR145 pKa = 11.84GGTWDD150 pKa = 3.99PPLANGDD157 pKa = 3.59RR158 pKa = 11.84GTFTIGTDD166 pKa = 3.06TPGIYY171 pKa = 9.35RR172 pKa = 11.84YY173 pKa = 8.8MISATAPCSDD183 pKa = 3.44SEE185 pKa = 4.41SRR187 pKa = 11.84VVVKK191 pKa = 10.55EE192 pKa = 4.01CFNDD196 pKa = 4.2DD197 pKa = 3.63FDD199 pKa = 6.03NDD201 pKa = 3.81GVNNKK206 pKa = 9.96DD207 pKa = 5.79DD208 pKa = 5.77LDD210 pKa = 5.14DD211 pKa = 5.77DD212 pKa = 4.37NDD214 pKa = 4.97GILDD218 pKa = 3.76SVEE221 pKa = 4.59NSACTSATLTEE232 pKa = 4.34SSPVVDD238 pKa = 3.69IDD240 pKa = 3.88FGTGNTPTTDD250 pKa = 3.64PNIQGHH256 pKa = 4.65QHH258 pKa = 5.51NPNWPDD264 pKa = 3.18DD265 pKa = 3.69GFYY268 pKa = 11.22NVANSLYY275 pKa = 9.25MFNSANFDD283 pKa = 3.26VWFITTDD290 pKa = 3.6TNPVPHH296 pKa = 7.05SDD298 pKa = 3.72GNGDD302 pKa = 3.4VNGRR306 pKa = 11.84FLAVNIASNFGNKK319 pKa = 7.89VLYY322 pKa = 9.05EE323 pKa = 4.18LKK325 pKa = 10.41DD326 pKa = 3.4IPISAGIQYY335 pKa = 10.08NFRR338 pKa = 11.84IDD340 pKa = 3.68MVGLCDD346 pKa = 4.59RR347 pKa = 11.84GCADD351 pKa = 4.29VPALDD356 pKa = 5.7LEE358 pKa = 5.61IIDD361 pKa = 3.78QTTGNTILSTTSAALGVANDD381 pKa = 4.76DD382 pKa = 2.82IWRR385 pKa = 11.84TLLDD389 pKa = 5.53DD390 pKa = 3.37ITVPTNTLLTLRR402 pKa = 11.84IRR404 pKa = 11.84NQQLQGSNGNDD415 pKa = 2.72IGIDD419 pKa = 3.59NIRR422 pKa = 11.84FAPLEE427 pKa = 4.35CDD429 pKa = 3.14SDD431 pKa = 4.25RR432 pKa = 11.84DD433 pKa = 3.8EE434 pKa = 4.83VPNFLDD440 pKa = 4.93LDD442 pKa = 4.27CDD444 pKa = 3.47NDD446 pKa = 4.78GIFDD450 pKa = 4.34IIEE453 pKa = 4.33AGRR456 pKa = 11.84SDD458 pKa = 5.5LDD460 pKa = 3.92TNGDD464 pKa = 3.96GIVDD468 pKa = 5.01GIRR471 pKa = 11.84DD472 pKa = 3.69TDD474 pKa = 3.48GDD476 pKa = 4.2GIADD480 pKa = 3.42VGVDD484 pKa = 3.66VNGVPIAASGGLTPISSDD502 pKa = 3.16GDD504 pKa = 3.84GLPDD508 pKa = 4.19YY509 pKa = 11.35LDD511 pKa = 3.91IDD513 pKa = 4.42ADD515 pKa = 3.73NDD517 pKa = 4.65GIQDD521 pKa = 4.06NIEE524 pKa = 4.07GQATLTYY531 pKa = 9.9IAPSGNDD538 pKa = 3.33LNKK541 pKa = 10.79NGVDD545 pKa = 3.58DD546 pKa = 4.99AYY548 pKa = 10.32EE549 pKa = 4.42GVGSITPVNTDD560 pKa = 2.75TDD562 pKa = 4.08TLPDD566 pKa = 3.97YY567 pKa = 11.49LDD569 pKa = 4.86LNSDD573 pKa = 4.17DD574 pKa = 5.57DD575 pKa = 4.32CLNDD579 pKa = 3.6TVEE582 pKa = 5.45AYY584 pKa = 10.25DD585 pKa = 4.56LNLDD589 pKa = 4.49GISDD593 pKa = 3.75ITAIGTDD600 pKa = 3.46TDD602 pKa = 3.7NDD604 pKa = 3.76GMDD607 pKa = 3.24NAFDD611 pKa = 4.26IVILDD616 pKa = 4.0ALTAITNPTDD626 pKa = 3.49NGEE629 pKa = 4.45LPTDD633 pKa = 4.18LPNNHH638 pKa = 6.85NIGGDD643 pKa = 3.06VDD645 pKa = 4.2FRR647 pKa = 11.84EE648 pKa = 4.24EE649 pKa = 3.91FKK651 pKa = 11.16KK652 pKa = 10.84VDD654 pKa = 3.6EE655 pKa = 4.78KK656 pKa = 10.79KK657 pKa = 8.85TAGGCASGIPTINLFDD673 pKa = 4.2SLVDD677 pKa = 3.48AAIPGGTWSGPSVLTGGEE695 pKa = 4.06SGTFNATTNVAGTYY709 pKa = 9.14TYY711 pKa = 9.84TLPTIGLCPSRR722 pKa = 11.84KK723 pKa = 9.92GEE725 pKa = 4.1VKK727 pKa = 10.56VSIISTSDD735 pKa = 2.81PGTNGTLNTCVLVTTPINLFDD756 pKa = 4.25SLGGTPDD763 pKa = 3.51TGGSWTDD770 pKa = 3.27PNGNPFGTTDD780 pKa = 3.3QGVLNPSATGTLQGTYY796 pKa = 9.83VYY798 pKa = 10.31TIGTTGCSSSANVEE812 pKa = 4.03VTINPGANAGLDD824 pKa = 3.53NKK826 pKa = 10.25VSFCSTQSSINLFDD840 pKa = 4.14SLEE843 pKa = 4.27GTPDD847 pKa = 3.33TGGSWIDD854 pKa = 3.42PNGNPFGTTDD864 pKa = 3.74QGVLDD869 pKa = 4.55PSLTTTLSGDD879 pKa = 3.57YY880 pKa = 9.66IYY882 pKa = 10.53TVSATTCPIQDD893 pKa = 3.62TAIVTVTIEE902 pKa = 3.79IAPNPGTNSSLEE914 pKa = 3.82ICTNASAINLFDD926 pKa = 4.06SLRR929 pKa = 11.84GAPTTGGTWTGPGGATFGTNDD950 pKa = 3.4KK951 pKa = 11.69GEE953 pKa = 4.42FNPASDD959 pKa = 3.61AAGTYY964 pKa = 9.19TYY966 pKa = 7.58TTGTIACNNTATITVTINPAANAGEE991 pKa = 4.11DD992 pKa = 3.61NKK994 pKa = 10.37VSFCSTDD1001 pKa = 2.7TTMNLFDD1008 pKa = 4.75SLGGSPNIGGSWTDD1022 pKa = 3.26PNGNPFGTTDD1032 pKa = 3.45QGIIDD1037 pKa = 4.45PSATTTLSGDD1047 pKa = 3.32YY1048 pKa = 9.54TYY1050 pKa = 10.8TVSTGKK1056 pKa = 8.55CTIPDD1061 pKa = 3.29IAKK1064 pKa = 7.59ITVVIEE1070 pKa = 4.03TTPSISFDD1078 pKa = 3.12KK1079 pKa = 8.29TTCSDD1084 pKa = 3.34DD1085 pKa = 3.72RR1086 pKa = 11.84NSYY1089 pKa = 10.4SVSFTSNEE1097 pKa = 3.83TWSITIDD1104 pKa = 3.63PPNAGIIDD1112 pKa = 3.92VTNSMITGIASDD1124 pKa = 3.74TNIIITATNPANTTCEE1140 pKa = 3.87DD1141 pKa = 3.96TLNIIAPDD1149 pKa = 4.03CTCPDD1154 pKa = 2.81ITEE1157 pKa = 4.25PTNPIHH1163 pKa = 7.07QRR1165 pKa = 11.84ICAGDD1170 pKa = 3.73TNPVLSVTVLTDD1182 pKa = 3.2QTANWYY1188 pKa = 7.21TQDD1191 pKa = 3.12GTLLVANTTTYY1202 pKa = 10.7TPIEE1206 pKa = 4.25TAIGTYY1212 pKa = 8.94TYY1214 pKa = 10.94SVEE1217 pKa = 4.95AFDD1220 pKa = 5.58NNEE1223 pKa = 3.91KK1224 pKa = 10.66CPSDD1228 pKa = 4.58RR1229 pKa = 11.84IPVFFDD1235 pKa = 2.65IVTAPEE1241 pKa = 3.8ITLSSDD1247 pKa = 3.25SAICVSEE1254 pKa = 4.92DD1255 pKa = 3.7GVPLPTNDD1263 pKa = 3.96FPFIDD1268 pKa = 3.89TFLNTTDD1275 pKa = 4.5FTFIWSFEE1283 pKa = 4.02GTVIPGQNSSGIEE1296 pKa = 3.83TLLPGTYY1303 pKa = 8.62TVQYY1307 pKa = 8.01TDD1309 pKa = 3.73KK1310 pKa = 11.33VSMCSDD1316 pKa = 3.62TSSVTINAIEE1326 pKa = 4.67GPQRR1330 pKa = 11.84LDD1332 pKa = 3.16LSLSLGTFADD1342 pKa = 3.87HH1343 pKa = 7.04NDD1345 pKa = 3.43IIATVAGNSEE1355 pKa = 4.53YY1356 pKa = 10.75EE1357 pKa = 3.9FSLDD1361 pKa = 3.2NGSFQTEE1368 pKa = 3.95NIFRR1372 pKa = 11.84NVALGLHH1379 pKa = 5.99EE1380 pKa = 4.24VTVRR1384 pKa = 11.84DD1385 pKa = 3.84IYY1387 pKa = 11.61GCGNLSKK1394 pKa = 10.3EE1395 pKa = 4.08IFVMGFPNFFTPNNDD1410 pKa = 4.37GINDD1414 pKa = 3.78TWNVIADD1421 pKa = 3.8EE1422 pKa = 4.8NLPEE1426 pKa = 4.0MIIYY1430 pKa = 10.32IFDD1433 pKa = 3.74RR1434 pKa = 11.84YY1435 pKa = 10.59GKK1437 pKa = 10.18LLSQLNPNGPGWDD1450 pKa = 3.74GIYY1453 pKa = 10.46NGRR1456 pKa = 11.84TLPSTDD1462 pKa = 2.89YY1463 pKa = 10.76WFAAQFKK1470 pKa = 9.87DD1471 pKa = 3.51GSEE1474 pKa = 4.42TYY1476 pKa = 10.07RR1477 pKa = 11.84SHH1479 pKa = 6.77FTLKK1483 pKa = 10.44RR1484 pKa = 3.41
MM1 pKa = 7.3NKK3 pKa = 8.26KK4 pKa = 8.54TYY6 pKa = 10.25LIVVILLTISKK17 pKa = 10.41GLFAQTMTLTSNAPSNTICRR37 pKa = 11.84GDD39 pKa = 3.85IIQVDD44 pKa = 4.21VNITPPNGASGYY56 pKa = 9.43NWTNVTTGQFIKK68 pKa = 10.62SVNGNAGTFHH78 pKa = 7.52LFTGVDD84 pKa = 3.27RR85 pKa = 11.84LLTNSTIRR93 pKa = 11.84VEE95 pKa = 4.4AFGGTGGNLSQTIDD109 pKa = 3.85IIVQQPANPGTEE121 pKa = 4.24GNLLLCNKK129 pKa = 9.35TGDD132 pKa = 3.39IDD134 pKa = 6.3LFTLLQGNPDD144 pKa = 4.2RR145 pKa = 11.84GGTWDD150 pKa = 3.99PPLANGDD157 pKa = 3.59RR158 pKa = 11.84GTFTIGTDD166 pKa = 3.06TPGIYY171 pKa = 9.35RR172 pKa = 11.84YY173 pKa = 8.8MISATAPCSDD183 pKa = 3.44SEE185 pKa = 4.41SRR187 pKa = 11.84VVVKK191 pKa = 10.55EE192 pKa = 4.01CFNDD196 pKa = 4.2DD197 pKa = 3.63FDD199 pKa = 6.03NDD201 pKa = 3.81GVNNKK206 pKa = 9.96DD207 pKa = 5.79DD208 pKa = 5.77LDD210 pKa = 5.14DD211 pKa = 5.77DD212 pKa = 4.37NDD214 pKa = 4.97GILDD218 pKa = 3.76SVEE221 pKa = 4.59NSACTSATLTEE232 pKa = 4.34SSPVVDD238 pKa = 3.69IDD240 pKa = 3.88FGTGNTPTTDD250 pKa = 3.64PNIQGHH256 pKa = 4.65QHH258 pKa = 5.51NPNWPDD264 pKa = 3.18DD265 pKa = 3.69GFYY268 pKa = 11.22NVANSLYY275 pKa = 9.25MFNSANFDD283 pKa = 3.26VWFITTDD290 pKa = 3.6TNPVPHH296 pKa = 7.05SDD298 pKa = 3.72GNGDD302 pKa = 3.4VNGRR306 pKa = 11.84FLAVNIASNFGNKK319 pKa = 7.89VLYY322 pKa = 9.05EE323 pKa = 4.18LKK325 pKa = 10.41DD326 pKa = 3.4IPISAGIQYY335 pKa = 10.08NFRR338 pKa = 11.84IDD340 pKa = 3.68MVGLCDD346 pKa = 4.59RR347 pKa = 11.84GCADD351 pKa = 4.29VPALDD356 pKa = 5.7LEE358 pKa = 5.61IIDD361 pKa = 3.78QTTGNTILSTTSAALGVANDD381 pKa = 4.76DD382 pKa = 2.82IWRR385 pKa = 11.84TLLDD389 pKa = 5.53DD390 pKa = 3.37ITVPTNTLLTLRR402 pKa = 11.84IRR404 pKa = 11.84NQQLQGSNGNDD415 pKa = 2.72IGIDD419 pKa = 3.59NIRR422 pKa = 11.84FAPLEE427 pKa = 4.35CDD429 pKa = 3.14SDD431 pKa = 4.25RR432 pKa = 11.84DD433 pKa = 3.8EE434 pKa = 4.83VPNFLDD440 pKa = 4.93LDD442 pKa = 4.27CDD444 pKa = 3.47NDD446 pKa = 4.78GIFDD450 pKa = 4.34IIEE453 pKa = 4.33AGRR456 pKa = 11.84SDD458 pKa = 5.5LDD460 pKa = 3.92TNGDD464 pKa = 3.96GIVDD468 pKa = 5.01GIRR471 pKa = 11.84DD472 pKa = 3.69TDD474 pKa = 3.48GDD476 pKa = 4.2GIADD480 pKa = 3.42VGVDD484 pKa = 3.66VNGVPIAASGGLTPISSDD502 pKa = 3.16GDD504 pKa = 3.84GLPDD508 pKa = 4.19YY509 pKa = 11.35LDD511 pKa = 3.91IDD513 pKa = 4.42ADD515 pKa = 3.73NDD517 pKa = 4.65GIQDD521 pKa = 4.06NIEE524 pKa = 4.07GQATLTYY531 pKa = 9.9IAPSGNDD538 pKa = 3.33LNKK541 pKa = 10.79NGVDD545 pKa = 3.58DD546 pKa = 4.99AYY548 pKa = 10.32EE549 pKa = 4.42GVGSITPVNTDD560 pKa = 2.75TDD562 pKa = 4.08TLPDD566 pKa = 3.97YY567 pKa = 11.49LDD569 pKa = 4.86LNSDD573 pKa = 4.17DD574 pKa = 5.57DD575 pKa = 4.32CLNDD579 pKa = 3.6TVEE582 pKa = 5.45AYY584 pKa = 10.25DD585 pKa = 4.56LNLDD589 pKa = 4.49GISDD593 pKa = 3.75ITAIGTDD600 pKa = 3.46TDD602 pKa = 3.7NDD604 pKa = 3.76GMDD607 pKa = 3.24NAFDD611 pKa = 4.26IVILDD616 pKa = 4.0ALTAITNPTDD626 pKa = 3.49NGEE629 pKa = 4.45LPTDD633 pKa = 4.18LPNNHH638 pKa = 6.85NIGGDD643 pKa = 3.06VDD645 pKa = 4.2FRR647 pKa = 11.84EE648 pKa = 4.24EE649 pKa = 3.91FKK651 pKa = 11.16KK652 pKa = 10.84VDD654 pKa = 3.6EE655 pKa = 4.78KK656 pKa = 10.79KK657 pKa = 8.85TAGGCASGIPTINLFDD673 pKa = 4.2SLVDD677 pKa = 3.48AAIPGGTWSGPSVLTGGEE695 pKa = 4.06SGTFNATTNVAGTYY709 pKa = 9.14TYY711 pKa = 9.84TLPTIGLCPSRR722 pKa = 11.84KK723 pKa = 9.92GEE725 pKa = 4.1VKK727 pKa = 10.56VSIISTSDD735 pKa = 2.81PGTNGTLNTCVLVTTPINLFDD756 pKa = 4.25SLGGTPDD763 pKa = 3.51TGGSWTDD770 pKa = 3.27PNGNPFGTTDD780 pKa = 3.3QGVLNPSATGTLQGTYY796 pKa = 9.83VYY798 pKa = 10.31TIGTTGCSSSANVEE812 pKa = 4.03VTINPGANAGLDD824 pKa = 3.53NKK826 pKa = 10.25VSFCSTQSSINLFDD840 pKa = 4.14SLEE843 pKa = 4.27GTPDD847 pKa = 3.33TGGSWIDD854 pKa = 3.42PNGNPFGTTDD864 pKa = 3.74QGVLDD869 pKa = 4.55PSLTTTLSGDD879 pKa = 3.57YY880 pKa = 9.66IYY882 pKa = 10.53TVSATTCPIQDD893 pKa = 3.62TAIVTVTIEE902 pKa = 3.79IAPNPGTNSSLEE914 pKa = 3.82ICTNASAINLFDD926 pKa = 4.06SLRR929 pKa = 11.84GAPTTGGTWTGPGGATFGTNDD950 pKa = 3.4KK951 pKa = 11.69GEE953 pKa = 4.42FNPASDD959 pKa = 3.61AAGTYY964 pKa = 9.19TYY966 pKa = 7.58TTGTIACNNTATITVTINPAANAGEE991 pKa = 4.11DD992 pKa = 3.61NKK994 pKa = 10.37VSFCSTDD1001 pKa = 2.7TTMNLFDD1008 pKa = 4.75SLGGSPNIGGSWTDD1022 pKa = 3.26PNGNPFGTTDD1032 pKa = 3.45QGIIDD1037 pKa = 4.45PSATTTLSGDD1047 pKa = 3.32YY1048 pKa = 9.54TYY1050 pKa = 10.8TVSTGKK1056 pKa = 8.55CTIPDD1061 pKa = 3.29IAKK1064 pKa = 7.59ITVVIEE1070 pKa = 4.03TTPSISFDD1078 pKa = 3.12KK1079 pKa = 8.29TTCSDD1084 pKa = 3.34DD1085 pKa = 3.72RR1086 pKa = 11.84NSYY1089 pKa = 10.4SVSFTSNEE1097 pKa = 3.83TWSITIDD1104 pKa = 3.63PPNAGIIDD1112 pKa = 3.92VTNSMITGIASDD1124 pKa = 3.74TNIIITATNPANTTCEE1140 pKa = 3.87DD1141 pKa = 3.96TLNIIAPDD1149 pKa = 4.03CTCPDD1154 pKa = 2.81ITEE1157 pKa = 4.25PTNPIHH1163 pKa = 7.07QRR1165 pKa = 11.84ICAGDD1170 pKa = 3.73TNPVLSVTVLTDD1182 pKa = 3.2QTANWYY1188 pKa = 7.21TQDD1191 pKa = 3.12GTLLVANTTTYY1202 pKa = 10.7TPIEE1206 pKa = 4.25TAIGTYY1212 pKa = 8.94TYY1214 pKa = 10.94SVEE1217 pKa = 4.95AFDD1220 pKa = 5.58NNEE1223 pKa = 3.91KK1224 pKa = 10.66CPSDD1228 pKa = 4.58RR1229 pKa = 11.84IPVFFDD1235 pKa = 2.65IVTAPEE1241 pKa = 3.8ITLSSDD1247 pKa = 3.25SAICVSEE1254 pKa = 4.92DD1255 pKa = 3.7GVPLPTNDD1263 pKa = 3.96FPFIDD1268 pKa = 3.89TFLNTTDD1275 pKa = 4.5FTFIWSFEE1283 pKa = 4.02GTVIPGQNSSGIEE1296 pKa = 3.83TLLPGTYY1303 pKa = 8.62TVQYY1307 pKa = 8.01TDD1309 pKa = 3.73KK1310 pKa = 11.33VSMCSDD1316 pKa = 3.62TSSVTINAIEE1326 pKa = 4.67GPQRR1330 pKa = 11.84LDD1332 pKa = 3.16LSLSLGTFADD1342 pKa = 3.87HH1343 pKa = 7.04NDD1345 pKa = 3.43IIATVAGNSEE1355 pKa = 4.53YY1356 pKa = 10.75EE1357 pKa = 3.9FSLDD1361 pKa = 3.2NGSFQTEE1368 pKa = 3.95NIFRR1372 pKa = 11.84NVALGLHH1379 pKa = 5.99EE1380 pKa = 4.24VTVRR1384 pKa = 11.84DD1385 pKa = 3.84IYY1387 pKa = 11.61GCGNLSKK1394 pKa = 10.3EE1395 pKa = 4.08IFVMGFPNFFTPNNDD1410 pKa = 4.37GINDD1414 pKa = 3.78TWNVIADD1421 pKa = 3.8EE1422 pKa = 4.8NLPEE1426 pKa = 4.0MIIYY1430 pKa = 10.32IFDD1433 pKa = 3.74RR1434 pKa = 11.84YY1435 pKa = 10.59GKK1437 pKa = 10.18LLSQLNPNGPGWDD1450 pKa = 3.74GIYY1453 pKa = 10.46NGRR1456 pKa = 11.84TLPSTDD1462 pKa = 2.89YY1463 pKa = 10.76WFAAQFKK1470 pKa = 9.87DD1471 pKa = 3.51GSEE1474 pKa = 4.42TYY1476 pKa = 10.07RR1477 pKa = 11.84SHH1479 pKa = 6.77FTLKK1483 pKa = 10.44RR1484 pKa = 3.41
Molecular weight: 157.34 kDa
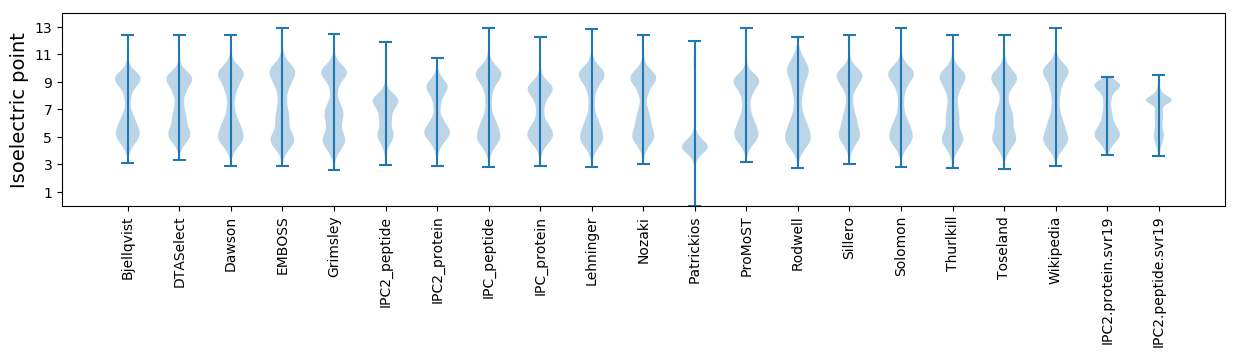
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A554VF87|A0A554VF87_9FLAO Uncharacterized protein OS=Aquimarina algiphila OX=2047982 GN=FOF46_21475 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
Molecular weight: 6.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2018512 |
22 |
4042 |
332.9 |
37.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.706 ± 0.032 | 0.742 ± 0.011 |
5.737 ± 0.027 | 6.589 ± 0.031 |
5.153 ± 0.027 | 6.308 ± 0.034 |
1.78 ± 0.015 | 8.466 ± 0.028 |
7.899 ± 0.044 | 8.982 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.014 ± 0.016 | 6.332 ± 0.032 |
3.351 ± 0.021 | 3.423 ± 0.018 |
3.508 ± 0.021 | 6.691 ± 0.024 |
6.161 ± 0.043 | 5.855 ± 0.022 |
1.089 ± 0.012 | 4.214 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
