
delta proteobacterium NaphS2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; unclassified Deltaproteobacteria
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
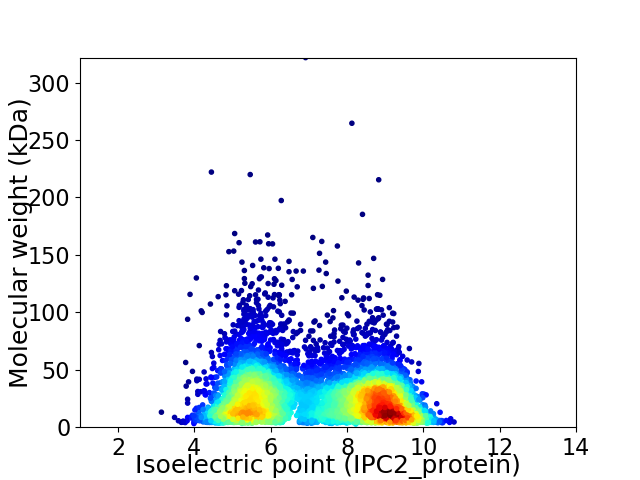
Virtual 2D-PAGE plot for 6687 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8F3A8|D8F3A8_9DELT Dehydrogenase FMN-dependent OS=delta proteobacterium NaphS2 OX=88274 GN=NPH_3944 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.05RR3 pKa = 11.84TTSVPPTYY11 pKa = 7.69TTKK14 pKa = 10.95ASNVSTALAGYY25 pKa = 8.72FAGGTGGAQSIDD37 pKa = 3.48TSEE40 pKa = 4.49GSPSVNSWKK49 pKa = 10.77GAGFQKK55 pKa = 10.09DD56 pKa = 3.35TTYY59 pKa = 10.38TANTAPYY66 pKa = 9.39FQITVDD72 pKa = 3.32TSKK75 pKa = 9.7YY76 pKa = 7.29TDD78 pKa = 3.46VIISLRR84 pKa = 11.84SYY86 pKa = 9.73ATANWGTSNVLKK98 pKa = 10.41VWSSADD104 pKa = 3.05GGTSFTATTPATGTLTKK121 pKa = 8.09TTWSGDD127 pKa = 3.4QTFAAAATGAVTTIFRR143 pKa = 11.84INAEE147 pKa = 4.09GANPTNGAAMMLDD160 pKa = 3.9NILVTGCGVPTPPPTITKK178 pKa = 10.06SFSPDD183 pKa = 3.14PTIKK187 pKa = 10.75GGTSTLTFTINNTEE201 pKa = 4.09SGNQALSGVAFTDD214 pKa = 3.54VLPEE218 pKa = 4.06GLFVADD224 pKa = 4.68DD225 pKa = 3.98TSPQCGGTLTTTAATRR241 pKa = 11.84TIALTGGTLAAGGNCTFDD259 pKa = 3.34VSVTGTLEE267 pKa = 3.92GQYY270 pKa = 10.57EE271 pKa = 4.24NVSGFISSNEE281 pKa = 4.23SGTSTHH287 pKa = 6.38YY288 pKa = 10.27ATDD291 pKa = 3.78SLNVIAPSILAKK303 pKa = 10.53SFAPASLLTGGSSTLTFSLTNPNQSGTLSGIGFTDD338 pKa = 3.92TLPAGLTVASGGPTAVCDD356 pKa = 3.91GTLTTTSPDD365 pKa = 3.59SISFSGGSLAANTTCTFDD383 pKa = 3.6VPVSGVTSGSKK394 pKa = 10.89ANTTGIITSTEE405 pKa = 3.93GGNGSSADD413 pKa = 3.31ATLNVSDD420 pKa = 4.39PVALIGLNKK429 pKa = 9.89EE430 pKa = 3.96VSTDD434 pKa = 3.02GTHH437 pKa = 6.02WYY439 pKa = 10.1KK440 pKa = 10.73FVGVPVGGNVYY451 pKa = 10.62YY452 pKa = 10.87RR453 pKa = 11.84FTVFNDD459 pKa = 4.03GEE461 pKa = 4.43ATLSSIDD468 pKa = 3.66VADD471 pKa = 4.28PDD473 pKa = 4.18ISLSSCSFPSSLAAGEE489 pKa = 4.51NPVSCTVGPITAVSGLHH506 pKa = 7.11DD507 pKa = 3.89NTATATGTYY516 pKa = 10.33GSQTPSDD523 pKa = 3.75TSTARR528 pKa = 11.84YY529 pKa = 9.42GSAEE533 pKa = 3.85LTIAKK538 pKa = 9.89SVTEE542 pKa = 3.9NDD544 pKa = 3.07FTTVGDD550 pKa = 3.87VLHH553 pKa = 6.18YY554 pKa = 10.66SYY556 pKa = 11.45LVTNSGSAPLLGPVTVSDD574 pKa = 4.48DD575 pKa = 3.6KK576 pKa = 12.09ANDD579 pKa = 3.77EE580 pKa = 4.35YY581 pKa = 11.02CPPVSTVGDD590 pKa = 3.89LDD592 pKa = 4.23NYY594 pKa = 10.47LDD596 pKa = 4.22PGEE599 pKa = 5.59SITCTAMYY607 pKa = 9.27TIEE610 pKa = 4.46SSDD613 pKa = 3.37IASGSVTNTATATADD628 pKa = 3.34GVTSNVDD635 pKa = 3.29SKK637 pKa = 10.06TVTSEE642 pKa = 4.16SIADD646 pKa = 3.64LSITKK651 pKa = 10.31SSTPDD656 pKa = 3.25PYY658 pKa = 11.52VPGDD662 pKa = 3.4GFTYY666 pKa = 9.96TIVVSNAGPSDD677 pKa = 3.58VTDD680 pKa = 4.04ARR682 pKa = 11.84VQDD685 pKa = 4.41ALPAALSGFTWTCTPNGVGAVCGSGSGTGDD715 pKa = 2.51IDD717 pKa = 4.72AFVTLPSGTSAIFSVSGTVPSGTTGDD743 pKa = 4.03LVNTATVQPPAGITDD758 pKa = 4.29PVPGSNSDD766 pKa = 3.18SDD768 pKa = 4.3TNPSGSAVSDD778 pKa = 3.5LSITKK783 pKa = 10.31SSTPDD788 pKa = 3.25PYY790 pKa = 11.52VPGDD794 pKa = 3.4GFTYY798 pKa = 9.96TIVVSNAGPSDD809 pKa = 3.58VTDD812 pKa = 4.04ARR814 pKa = 11.84VQDD817 pKa = 4.41ALPAALSGFAWTCTSNGGGAVCGSAGGTGDD847 pKa = 2.79IDD849 pKa = 4.33AFVSLPAGTSVNFSVTGTVPSGTTGSLMNTATIQPPSGTTDD890 pKa = 3.07PVTGNNSDD898 pKa = 3.64SDD900 pKa = 4.3TNPGGGPAIPTTNEE914 pKa = 3.27YY915 pKa = 11.35GLIFLAILLVLFSLNHH931 pKa = 6.68LGRR934 pKa = 11.84QRR936 pKa = 11.84RR937 pKa = 11.84II938 pKa = 3.29
MM1 pKa = 7.44KK2 pKa = 9.05RR3 pKa = 11.84TTSVPPTYY11 pKa = 7.69TTKK14 pKa = 10.95ASNVSTALAGYY25 pKa = 8.72FAGGTGGAQSIDD37 pKa = 3.48TSEE40 pKa = 4.49GSPSVNSWKK49 pKa = 10.77GAGFQKK55 pKa = 10.09DD56 pKa = 3.35TTYY59 pKa = 10.38TANTAPYY66 pKa = 9.39FQITVDD72 pKa = 3.32TSKK75 pKa = 9.7YY76 pKa = 7.29TDD78 pKa = 3.46VIISLRR84 pKa = 11.84SYY86 pKa = 9.73ATANWGTSNVLKK98 pKa = 10.41VWSSADD104 pKa = 3.05GGTSFTATTPATGTLTKK121 pKa = 8.09TTWSGDD127 pKa = 3.4QTFAAAATGAVTTIFRR143 pKa = 11.84INAEE147 pKa = 4.09GANPTNGAAMMLDD160 pKa = 3.9NILVTGCGVPTPPPTITKK178 pKa = 10.06SFSPDD183 pKa = 3.14PTIKK187 pKa = 10.75GGTSTLTFTINNTEE201 pKa = 4.09SGNQALSGVAFTDD214 pKa = 3.54VLPEE218 pKa = 4.06GLFVADD224 pKa = 4.68DD225 pKa = 3.98TSPQCGGTLTTTAATRR241 pKa = 11.84TIALTGGTLAAGGNCTFDD259 pKa = 3.34VSVTGTLEE267 pKa = 3.92GQYY270 pKa = 10.57EE271 pKa = 4.24NVSGFISSNEE281 pKa = 4.23SGTSTHH287 pKa = 6.38YY288 pKa = 10.27ATDD291 pKa = 3.78SLNVIAPSILAKK303 pKa = 10.53SFAPASLLTGGSSTLTFSLTNPNQSGTLSGIGFTDD338 pKa = 3.92TLPAGLTVASGGPTAVCDD356 pKa = 3.91GTLTTTSPDD365 pKa = 3.59SISFSGGSLAANTTCTFDD383 pKa = 3.6VPVSGVTSGSKK394 pKa = 10.89ANTTGIITSTEE405 pKa = 3.93GGNGSSADD413 pKa = 3.31ATLNVSDD420 pKa = 4.39PVALIGLNKK429 pKa = 9.89EE430 pKa = 3.96VSTDD434 pKa = 3.02GTHH437 pKa = 6.02WYY439 pKa = 10.1KK440 pKa = 10.73FVGVPVGGNVYY451 pKa = 10.62YY452 pKa = 10.87RR453 pKa = 11.84FTVFNDD459 pKa = 4.03GEE461 pKa = 4.43ATLSSIDD468 pKa = 3.66VADD471 pKa = 4.28PDD473 pKa = 4.18ISLSSCSFPSSLAAGEE489 pKa = 4.51NPVSCTVGPITAVSGLHH506 pKa = 7.11DD507 pKa = 3.89NTATATGTYY516 pKa = 10.33GSQTPSDD523 pKa = 3.75TSTARR528 pKa = 11.84YY529 pKa = 9.42GSAEE533 pKa = 3.85LTIAKK538 pKa = 9.89SVTEE542 pKa = 3.9NDD544 pKa = 3.07FTTVGDD550 pKa = 3.87VLHH553 pKa = 6.18YY554 pKa = 10.66SYY556 pKa = 11.45LVTNSGSAPLLGPVTVSDD574 pKa = 4.48DD575 pKa = 3.6KK576 pKa = 12.09ANDD579 pKa = 3.77EE580 pKa = 4.35YY581 pKa = 11.02CPPVSTVGDD590 pKa = 3.89LDD592 pKa = 4.23NYY594 pKa = 10.47LDD596 pKa = 4.22PGEE599 pKa = 5.59SITCTAMYY607 pKa = 9.27TIEE610 pKa = 4.46SSDD613 pKa = 3.37IASGSVTNTATATADD628 pKa = 3.34GVTSNVDD635 pKa = 3.29SKK637 pKa = 10.06TVTSEE642 pKa = 4.16SIADD646 pKa = 3.64LSITKK651 pKa = 10.31SSTPDD656 pKa = 3.25PYY658 pKa = 11.52VPGDD662 pKa = 3.4GFTYY666 pKa = 9.96TIVVSNAGPSDD677 pKa = 3.58VTDD680 pKa = 4.04ARR682 pKa = 11.84VQDD685 pKa = 4.41ALPAALSGFTWTCTPNGVGAVCGSGSGTGDD715 pKa = 2.51IDD717 pKa = 4.72AFVTLPSGTSAIFSVSGTVPSGTTGDD743 pKa = 4.03LVNTATVQPPAGITDD758 pKa = 4.29PVPGSNSDD766 pKa = 3.18SDD768 pKa = 4.3TNPSGSAVSDD778 pKa = 3.5LSITKK783 pKa = 10.31SSTPDD788 pKa = 3.25PYY790 pKa = 11.52VPGDD794 pKa = 3.4GFTYY798 pKa = 9.96TIVVSNAGPSDD809 pKa = 3.58VTDD812 pKa = 4.04ARR814 pKa = 11.84VQDD817 pKa = 4.41ALPAALSGFAWTCTSNGGGAVCGSAGGTGDD847 pKa = 2.79IDD849 pKa = 4.33AFVSLPAGTSVNFSVTGTVPSGTTGSLMNTATIQPPSGTTDD890 pKa = 3.07PVTGNNSDD898 pKa = 3.64SDD900 pKa = 4.3TNPGGGPAIPTTNEE914 pKa = 3.27YY915 pKa = 11.35GLIFLAILLVLFSLNHH931 pKa = 6.68LGRR934 pKa = 11.84QRR936 pKa = 11.84RR937 pKa = 11.84II938 pKa = 3.29
Molecular weight: 93.95 kDa
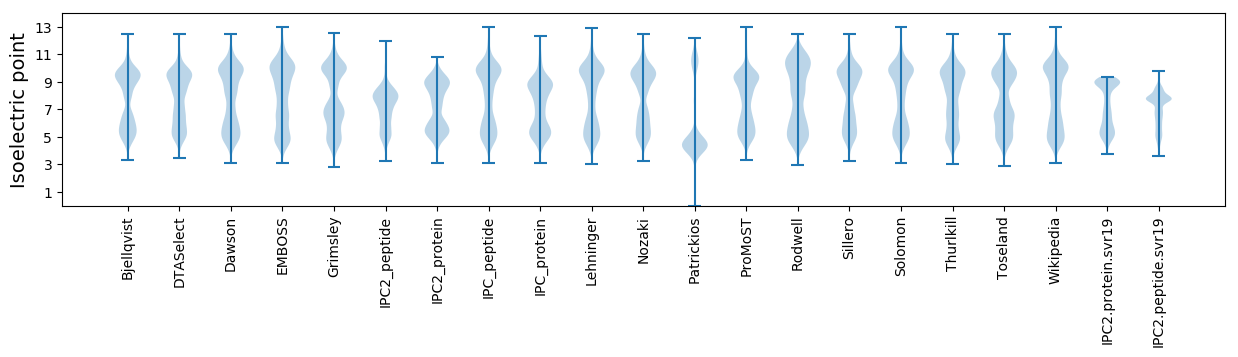
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8F2J8|D8F2J8_9DELT Uncharacterized protein OS=delta proteobacterium NaphS2 OX=88274 GN=NPH_0585 PE=4 SV=1
MM1 pKa = 7.4NVRR4 pKa = 11.84KK5 pKa = 10.0INAIKK10 pKa = 9.44HH11 pKa = 5.1QKK13 pKa = 9.3NKK15 pKa = 10.14SIPIFRR21 pKa = 11.84SNINAIVRR29 pKa = 11.84NAFVGKK35 pKa = 10.05RR36 pKa = 11.84AFKK39 pKa = 10.74
MM1 pKa = 7.4NVRR4 pKa = 11.84KK5 pKa = 10.0INAIKK10 pKa = 9.44HH11 pKa = 5.1QKK13 pKa = 9.3NKK15 pKa = 10.14SIPIFRR21 pKa = 11.84SNINAIVRR29 pKa = 11.84NAFVGKK35 pKa = 10.05RR36 pKa = 11.84AFKK39 pKa = 10.74
Molecular weight: 4.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1789710 |
30 |
2881 |
267.6 |
30.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.799 ± 0.03 | 1.366 ± 0.013 |
5.315 ± 0.023 | 6.659 ± 0.034 |
4.518 ± 0.023 | 7.426 ± 0.03 |
2.24 ± 0.015 | 6.442 ± 0.024 |
6.361 ± 0.03 | 9.97 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.83 ± 0.015 | 3.654 ± 0.02 |
4.58 ± 0.019 | 3.178 ± 0.019 |
6.204 ± 0.03 | 5.721 ± 0.021 |
4.85 ± 0.023 | 6.677 ± 0.025 |
1.204 ± 0.013 | 3.002 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
