
Deinbollia mosaic virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.3
Get precalculated fractions of proteins
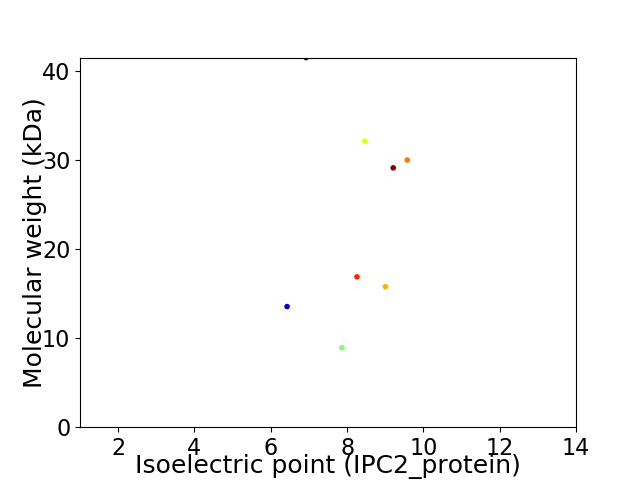
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

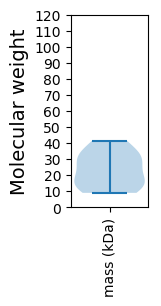
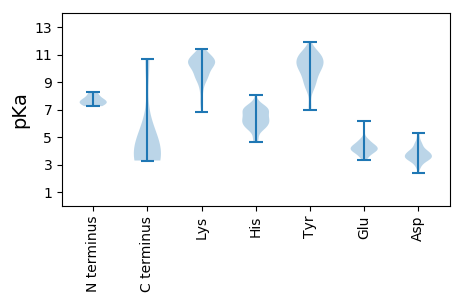
Protein with the lowest isoelectric point:
>tr|A0A142C6K7|A0A142C6K7_9GEMI Capsid protein OS=Deinbollia mosaic virus OX=1812308 GN=AV1 PE=3 SV=1
MM1 pKa = 7.99WDD3 pKa = 3.38PLVNEE8 pKa = 5.24FPDD11 pKa = 3.72SVHH14 pKa = 6.81GFRR17 pKa = 11.84CMLAIKK23 pKa = 9.31YY24 pKa = 8.48LQAIEE29 pKa = 4.11EE30 pKa = 4.56TYY32 pKa = 10.77EE33 pKa = 4.16PNTLGHH39 pKa = 6.99DD40 pKa = 5.01LIRR43 pKa = 11.84DD44 pKa = 4.58LISVIRR50 pKa = 11.84ARR52 pKa = 11.84DD53 pKa = 3.49YY54 pKa = 11.1VEE56 pKa = 3.35ATRR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 8.32NHH63 pKa = 5.68FHH65 pKa = 6.31SRR67 pKa = 11.84LEE69 pKa = 4.38GASKK73 pKa = 9.9TEE75 pKa = 3.89LRR77 pKa = 11.84QPICQPCCCPYY88 pKa = 10.45CPRR91 pKa = 11.84HH92 pKa = 5.76KK93 pKa = 9.6QTSSMDD99 pKa = 3.66LQAHH103 pKa = 4.65VQKK106 pKa = 10.96AQDD109 pKa = 3.55VQNVQKK115 pKa = 10.64PP116 pKa = 3.36
MM1 pKa = 7.99WDD3 pKa = 3.38PLVNEE8 pKa = 5.24FPDD11 pKa = 3.72SVHH14 pKa = 6.81GFRR17 pKa = 11.84CMLAIKK23 pKa = 9.31YY24 pKa = 8.48LQAIEE29 pKa = 4.11EE30 pKa = 4.56TYY32 pKa = 10.77EE33 pKa = 4.16PNTLGHH39 pKa = 6.99DD40 pKa = 5.01LIRR43 pKa = 11.84DD44 pKa = 4.58LISVIRR50 pKa = 11.84ARR52 pKa = 11.84DD53 pKa = 3.49YY54 pKa = 11.1VEE56 pKa = 3.35ATRR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 8.32NHH63 pKa = 5.68FHH65 pKa = 6.31SRR67 pKa = 11.84LEE69 pKa = 4.38GASKK73 pKa = 9.9TEE75 pKa = 3.89LRR77 pKa = 11.84QPICQPCCCPYY88 pKa = 10.45CPRR91 pKa = 11.84HH92 pKa = 5.76KK93 pKa = 9.6QTSSMDD99 pKa = 3.66LQAHH103 pKa = 4.65VQKK106 pKa = 10.96AQDD109 pKa = 3.55VQNVQKK115 pKa = 10.64PP116 pKa = 3.36
Molecular weight: 13.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142C6K8|A0A142C6K8_9GEMI AC4 OS=Deinbollia mosaic virus OX=1812308 GN=AC4 PE=3 SV=1
MM1 pKa = 7.7SKK3 pKa = 10.44RR4 pKa = 11.84PGDD7 pKa = 3.89IIISTPASKK16 pKa = 10.34VRR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 10.58LNFDD24 pKa = 3.17SPYY27 pKa = 9.22ASRR30 pKa = 11.84VAVPIVQGTNRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84AWTYY47 pKa = 9.4RR48 pKa = 11.84PMYY51 pKa = 9.33RR52 pKa = 11.84KK53 pKa = 9.49PKK55 pKa = 7.48MYY57 pKa = 10.84RR58 pKa = 11.84MFRR61 pKa = 11.84SLDD64 pKa = 3.4VPRR67 pKa = 11.84GCEE70 pKa = 4.36GPCKK74 pKa = 9.32VQSYY78 pKa = 6.99EE79 pKa = 3.67QRR81 pKa = 11.84DD82 pKa = 4.05DD83 pKa = 3.72VKK85 pKa = 9.5HH86 pKa = 5.48TGIVRR91 pKa = 11.84CVSDD95 pKa = 3.36VTRR98 pKa = 11.84GPGITHH104 pKa = 6.76RR105 pKa = 11.84VGKK108 pKa = 9.5RR109 pKa = 11.84FCVKK113 pKa = 10.3SIYY116 pKa = 10.16ILGKK120 pKa = 9.79VWMDD124 pKa = 4.63DD125 pKa = 3.63NIKK128 pKa = 10.13KK129 pKa = 9.97QNHH132 pKa = 4.98TNQVMFFLVRR142 pKa = 11.84DD143 pKa = 3.69RR144 pKa = 11.84RR145 pKa = 11.84PYY147 pKa = 8.77GTSPQDD153 pKa = 3.2FGQVFNMFDD162 pKa = 3.99NEE164 pKa = 4.11PSTATVKK171 pKa = 10.81NDD173 pKa = 2.84LRR175 pKa = 11.84DD176 pKa = 3.47RR177 pKa = 11.84YY178 pKa = 9.28QVLRR182 pKa = 11.84KK183 pKa = 8.77FHH185 pKa = 5.93ATVVGGPSGMKK196 pKa = 9.33EE197 pKa = 3.56QSLVKK202 pKa = 10.4RR203 pKa = 11.84FFRR206 pKa = 11.84VNSHH210 pKa = 4.87VVYY213 pKa = 10.59NHH215 pKa = 5.69QEE217 pKa = 3.45AAKK220 pKa = 10.4YY221 pKa = 9.25EE222 pKa = 4.13NHH224 pKa = 6.46TEE226 pKa = 3.99NALLLYY232 pKa = 7.29MACTHH237 pKa = 7.07ASNPVYY243 pKa = 9.86ATLKK247 pKa = 9.47IRR249 pKa = 11.84IYY251 pKa = 10.65FYY253 pKa = 11.26DD254 pKa = 3.34AVTNN258 pKa = 4.16
MM1 pKa = 7.7SKK3 pKa = 10.44RR4 pKa = 11.84PGDD7 pKa = 3.89IIISTPASKK16 pKa = 10.34VRR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 10.58LNFDD24 pKa = 3.17SPYY27 pKa = 9.22ASRR30 pKa = 11.84VAVPIVQGTNRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84AWTYY47 pKa = 9.4RR48 pKa = 11.84PMYY51 pKa = 9.33RR52 pKa = 11.84KK53 pKa = 9.49PKK55 pKa = 7.48MYY57 pKa = 10.84RR58 pKa = 11.84MFRR61 pKa = 11.84SLDD64 pKa = 3.4VPRR67 pKa = 11.84GCEE70 pKa = 4.36GPCKK74 pKa = 9.32VQSYY78 pKa = 6.99EE79 pKa = 3.67QRR81 pKa = 11.84DD82 pKa = 4.05DD83 pKa = 3.72VKK85 pKa = 9.5HH86 pKa = 5.48TGIVRR91 pKa = 11.84CVSDD95 pKa = 3.36VTRR98 pKa = 11.84GPGITHH104 pKa = 6.76RR105 pKa = 11.84VGKK108 pKa = 9.5RR109 pKa = 11.84FCVKK113 pKa = 10.3SIYY116 pKa = 10.16ILGKK120 pKa = 9.79VWMDD124 pKa = 4.63DD125 pKa = 3.63NIKK128 pKa = 10.13KK129 pKa = 9.97QNHH132 pKa = 4.98TNQVMFFLVRR142 pKa = 11.84DD143 pKa = 3.69RR144 pKa = 11.84RR145 pKa = 11.84PYY147 pKa = 8.77GTSPQDD153 pKa = 3.2FGQVFNMFDD162 pKa = 3.99NEE164 pKa = 4.11PSTATVKK171 pKa = 10.81NDD173 pKa = 2.84LRR175 pKa = 11.84DD176 pKa = 3.47RR177 pKa = 11.84YY178 pKa = 9.28QVLRR182 pKa = 11.84KK183 pKa = 8.77FHH185 pKa = 5.93ATVVGGPSGMKK196 pKa = 9.33EE197 pKa = 3.56QSLVKK202 pKa = 10.4RR203 pKa = 11.84FFRR206 pKa = 11.84VNSHH210 pKa = 4.87VVYY213 pKa = 10.59NHH215 pKa = 5.69QEE217 pKa = 3.45AAKK220 pKa = 10.4YY221 pKa = 9.25EE222 pKa = 4.13NHH224 pKa = 6.46TEE226 pKa = 3.99NALLLYY232 pKa = 7.29MACTHH237 pKa = 7.07ASNPVYY243 pKa = 9.86ATLKK247 pKa = 9.47IRR249 pKa = 11.84IYY251 pKa = 10.65FYY253 pKa = 11.26DD254 pKa = 3.34AVTNN258 pKa = 4.16
Molecular weight: 29.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1633 |
79 |
364 |
204.1 |
23.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.021 ± 0.589 | 2.082 ± 0.408 |
4.96 ± 0.237 | 4.409 ± 0.558 |
4.348 ± 0.317 | 4.838 ± 0.341 |
3.307 ± 0.464 | 6.43 ± 0.542 |
5.818 ± 0.539 | 6.981 ± 0.479 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.837 ± 0.419 | 6.369 ± 0.547 |
5.695 ± 0.441 | 5.45 ± 0.831 |
7.287 ± 0.694 | 8.634 ± 0.742 |
5.94 ± 0.49 | 5.205 ± 0.938 |
1.408 ± 0.189 | 3.98 ± 0.486 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
