
Hubei sobemo-like virus 32
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
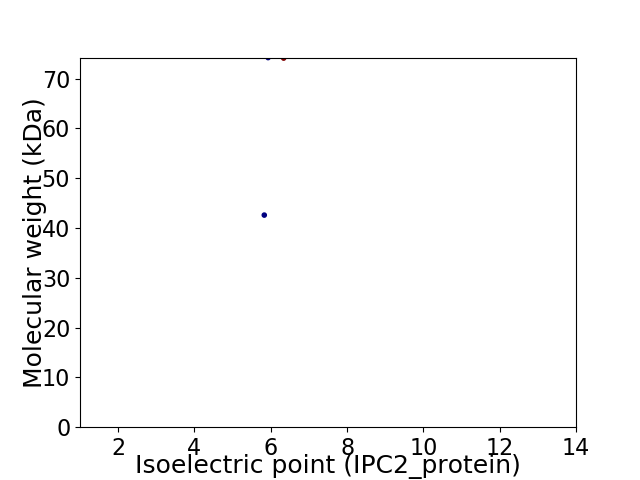
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEM3|A0A1L3KEM3_9VIRU C2H2-type domain-containing protein OS=Hubei sobemo-like virus 32 OX=1923219 PE=4 SV=1
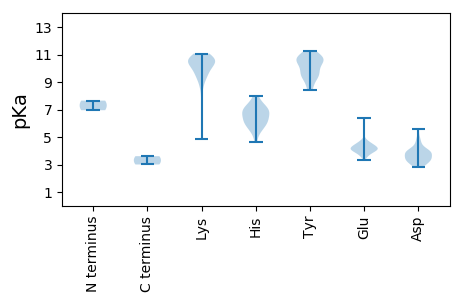
MM1 pKa = 6.95VCDD4 pKa = 4.85RR5 pKa = 11.84MVSCKK10 pKa = 7.71WRR12 pKa = 11.84IPSDD16 pKa = 3.39FLTRR20 pKa = 11.84GHH22 pKa = 6.8FDD24 pKa = 2.93RR25 pKa = 11.84VVRR28 pKa = 11.84EE29 pKa = 4.33LEE31 pKa = 4.17WTSSPGYY38 pKa = 8.44PYY40 pKa = 10.53CVRR43 pKa = 11.84HH44 pKa = 5.31PTNAQFFEE52 pKa = 4.63VVDD55 pKa = 4.27GQPSQIALDD64 pKa = 3.94RR65 pKa = 11.84VWYY68 pKa = 8.87MVVQRR73 pKa = 11.84LRR75 pKa = 11.84DD76 pKa = 3.58QTSDD80 pKa = 4.1PIRR83 pKa = 11.84LFIKK87 pKa = 10.6PEE89 pKa = 3.56AHH91 pKa = 7.07KK92 pKa = 10.15MSKK95 pKa = 10.29LLEE98 pKa = 3.8HH99 pKa = 7.75RR100 pKa = 11.84YY101 pKa = 10.07RR102 pKa = 11.84IISSVSVIDD111 pKa = 4.29QIIDD115 pKa = 3.14HH116 pKa = 6.33MLFQEE121 pKa = 4.22FNDD124 pKa = 3.24RR125 pKa = 11.84VVEE128 pKa = 4.12NVHH131 pKa = 5.62YY132 pKa = 10.95VPAKK136 pKa = 10.55VGWTPYY142 pKa = 9.5IGGWKK147 pKa = 9.33MIRR150 pKa = 11.84PHH152 pKa = 7.97GMVSADD158 pKa = 2.96KK159 pKa = 9.85TAWDD163 pKa = 3.46WTVPMWMLDD172 pKa = 2.85WEE174 pKa = 4.49FQVRR178 pKa = 11.84VKK180 pKa = 10.75LCEE183 pKa = 3.83NMTEE187 pKa = 3.83QWLDD191 pKa = 3.37LAAWRR196 pKa = 11.84YY197 pKa = 9.32NKK199 pKa = 10.39LFVDD203 pKa = 4.11VTLVTSGGLLLKK215 pKa = 10.36QRR217 pKa = 11.84LPGAMKK223 pKa = 10.19SGSVNTIVSNSIMQDD238 pKa = 2.88LMHH241 pKa = 7.19CYY243 pKa = 9.64ICNKK247 pKa = 9.84LDD249 pKa = 3.47IPVGHH254 pKa = 7.35IWTLGDD260 pKa = 3.73DD261 pKa = 5.05TIQEE265 pKa = 4.48DD266 pKa = 3.48PGEE269 pKa = 4.19EE270 pKa = 4.05YY271 pKa = 10.79FEE273 pKa = 4.16EE274 pKa = 4.28LKK276 pKa = 10.65KK277 pKa = 11.07LCVIKK282 pKa = 10.52EE283 pKa = 4.05IVRR286 pKa = 11.84GTEE289 pKa = 4.01FAGHH293 pKa = 6.35TFDD296 pKa = 5.61GMHH299 pKa = 6.55VSPNYY304 pKa = 9.58VGKK307 pKa = 9.4HH308 pKa = 5.69AYY310 pKa = 8.5TLLHH314 pKa = 6.03VDD316 pKa = 3.71LRR318 pKa = 11.84HH319 pKa = 6.05AKK321 pKa = 8.79EE322 pKa = 3.54ICMSYY327 pKa = 11.05ALLYY331 pKa = 10.62HH332 pKa = 6.76RR333 pKa = 11.84AKK335 pKa = 10.71PSDD338 pKa = 3.39KK339 pKa = 10.62EE340 pKa = 4.28LIYY343 pKa = 10.19TALRR347 pKa = 11.84SLGEE351 pKa = 4.18TLPSEE356 pKa = 4.47AKK358 pKa = 10.31LDD360 pKa = 3.83LVFDD364 pKa = 4.38GSEE367 pKa = 3.61
MM1 pKa = 6.95VCDD4 pKa = 4.85RR5 pKa = 11.84MVSCKK10 pKa = 7.71WRR12 pKa = 11.84IPSDD16 pKa = 3.39FLTRR20 pKa = 11.84GHH22 pKa = 6.8FDD24 pKa = 2.93RR25 pKa = 11.84VVRR28 pKa = 11.84EE29 pKa = 4.33LEE31 pKa = 4.17WTSSPGYY38 pKa = 8.44PYY40 pKa = 10.53CVRR43 pKa = 11.84HH44 pKa = 5.31PTNAQFFEE52 pKa = 4.63VVDD55 pKa = 4.27GQPSQIALDD64 pKa = 3.94RR65 pKa = 11.84VWYY68 pKa = 8.87MVVQRR73 pKa = 11.84LRR75 pKa = 11.84DD76 pKa = 3.58QTSDD80 pKa = 4.1PIRR83 pKa = 11.84LFIKK87 pKa = 10.6PEE89 pKa = 3.56AHH91 pKa = 7.07KK92 pKa = 10.15MSKK95 pKa = 10.29LLEE98 pKa = 3.8HH99 pKa = 7.75RR100 pKa = 11.84YY101 pKa = 10.07RR102 pKa = 11.84IISSVSVIDD111 pKa = 4.29QIIDD115 pKa = 3.14HH116 pKa = 6.33MLFQEE121 pKa = 4.22FNDD124 pKa = 3.24RR125 pKa = 11.84VVEE128 pKa = 4.12NVHH131 pKa = 5.62YY132 pKa = 10.95VPAKK136 pKa = 10.55VGWTPYY142 pKa = 9.5IGGWKK147 pKa = 9.33MIRR150 pKa = 11.84PHH152 pKa = 7.97GMVSADD158 pKa = 2.96KK159 pKa = 9.85TAWDD163 pKa = 3.46WTVPMWMLDD172 pKa = 2.85WEE174 pKa = 4.49FQVRR178 pKa = 11.84VKK180 pKa = 10.75LCEE183 pKa = 3.83NMTEE187 pKa = 3.83QWLDD191 pKa = 3.37LAAWRR196 pKa = 11.84YY197 pKa = 9.32NKK199 pKa = 10.39LFVDD203 pKa = 4.11VTLVTSGGLLLKK215 pKa = 10.36QRR217 pKa = 11.84LPGAMKK223 pKa = 10.19SGSVNTIVSNSIMQDD238 pKa = 2.88LMHH241 pKa = 7.19CYY243 pKa = 9.64ICNKK247 pKa = 9.84LDD249 pKa = 3.47IPVGHH254 pKa = 7.35IWTLGDD260 pKa = 3.73DD261 pKa = 5.05TIQEE265 pKa = 4.48DD266 pKa = 3.48PGEE269 pKa = 4.19EE270 pKa = 4.05YY271 pKa = 10.79FEE273 pKa = 4.16EE274 pKa = 4.28LKK276 pKa = 10.65KK277 pKa = 11.07LCVIKK282 pKa = 10.52EE283 pKa = 4.05IVRR286 pKa = 11.84GTEE289 pKa = 4.01FAGHH293 pKa = 6.35TFDD296 pKa = 5.61GMHH299 pKa = 6.55VSPNYY304 pKa = 9.58VGKK307 pKa = 9.4HH308 pKa = 5.69AYY310 pKa = 8.5TLLHH314 pKa = 6.03VDD316 pKa = 3.71LRR318 pKa = 11.84HH319 pKa = 6.05AKK321 pKa = 8.79EE322 pKa = 3.54ICMSYY327 pKa = 11.05ALLYY331 pKa = 10.62HH332 pKa = 6.76RR333 pKa = 11.84AKK335 pKa = 10.71PSDD338 pKa = 3.39KK339 pKa = 10.62EE340 pKa = 4.28LIYY343 pKa = 10.19TALRR347 pKa = 11.84SLGEE351 pKa = 4.18TLPSEE356 pKa = 4.47AKK358 pKa = 10.31LDD360 pKa = 3.83LVFDD364 pKa = 4.38GSEE367 pKa = 3.61
Molecular weight: 42.59 kDa
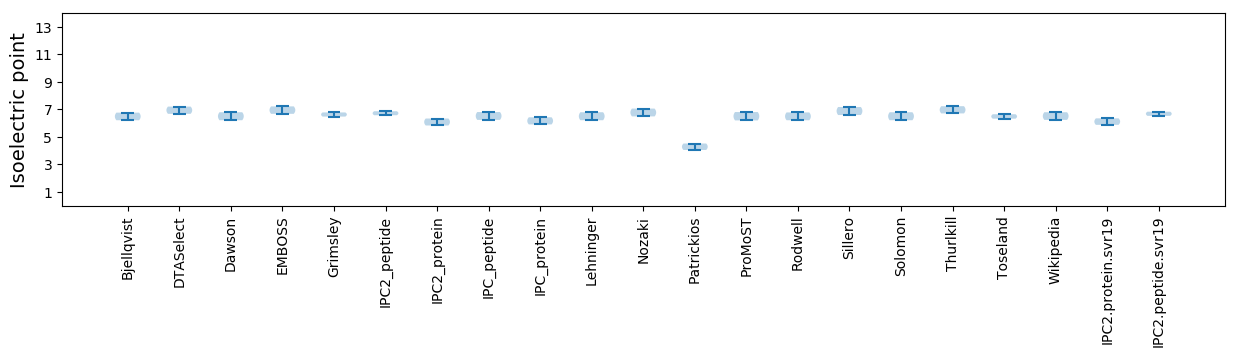
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEM3|A0A1L3KEM3_9VIRU C2H2-type domain-containing protein OS=Hubei sobemo-like virus 32 OX=1923219 PE=4 SV=1
MM1 pKa = 7.65EE2 pKa = 6.03FLRR5 pKa = 11.84QLTWFKK11 pKa = 10.24MAWLIAKK18 pKa = 9.55FYY20 pKa = 9.31MGFVMIMNISTILTFLVTLLVSLTWKK46 pKa = 9.39MIKK49 pKa = 9.94KK50 pKa = 9.57VALVDD55 pKa = 3.77LFYY58 pKa = 11.04EE59 pKa = 4.76GFGSVVDD66 pKa = 3.98GEE68 pKa = 4.78VIHH71 pKa = 6.03TVVEE75 pKa = 4.51AVRR78 pKa = 11.84EE79 pKa = 4.1AHH81 pKa = 6.19EE82 pKa = 4.07ATRR85 pKa = 11.84PVEE88 pKa = 4.1PPSWWEE94 pKa = 3.52EE95 pKa = 4.05AYY97 pKa = 10.66VWLMDD102 pKa = 3.52MAMDD106 pKa = 3.92NFDD109 pKa = 3.72VLRR112 pKa = 11.84ICLVMVAGLLVLLAVKK128 pKa = 10.33RR129 pKa = 11.84LVWRR133 pKa = 11.84RR134 pKa = 11.84AKK136 pKa = 10.53RR137 pKa = 11.84LVYY140 pKa = 9.48TMKK143 pKa = 10.65GFHH146 pKa = 6.86CEE148 pKa = 3.73AMVEE152 pKa = 4.29GSEE155 pKa = 4.45FGSGDD160 pKa = 3.16IPSFQIPMYY169 pKa = 9.8RR170 pKa = 11.84AGIFVSTFLGYY181 pKa = 9.87GVRR184 pKa = 11.84VDD186 pKa = 3.58NTLCVPTHH194 pKa = 5.4VVNLARR200 pKa = 11.84GGQLMADD207 pKa = 3.52TWILPRR213 pKa = 11.84TGILSKK219 pKa = 10.34LHH221 pKa = 6.95PDD223 pKa = 2.83VTYY226 pKa = 10.84YY227 pKa = 10.62YY228 pKa = 10.41LPEE231 pKa = 4.3KK232 pKa = 8.69TWSVLATSKK241 pKa = 10.78PKK243 pKa = 10.47VGPVMAAVVVSCAGPDD259 pKa = 4.01GVSHH263 pKa = 5.93GHH265 pKa = 6.78LRR267 pKa = 11.84QTPDD271 pKa = 3.38LGQVTYY277 pKa = 10.71SGSTLPGFSGAAYY290 pKa = 10.41FNGDD294 pKa = 3.89VLFGMHH300 pKa = 6.49TGVACEE306 pKa = 3.99RR307 pKa = 11.84NIGVAAHH314 pKa = 7.15LLQFEE319 pKa = 4.16ARR321 pKa = 11.84EE322 pKa = 4.1AFGQMEE328 pKa = 4.75YY329 pKa = 10.59KK330 pKa = 10.76GEE332 pKa = 4.15SPLNDD337 pKa = 3.32KK338 pKa = 8.61THH340 pKa = 7.12DD341 pKa = 3.9KK342 pKa = 10.83VLKK345 pKa = 7.08TQNRR349 pKa = 11.84VVTWKK354 pKa = 10.82SSDD357 pKa = 3.22ILEE360 pKa = 4.46KK361 pKa = 10.71AKK363 pKa = 10.76KK364 pKa = 9.4LWGPAGSSWADD375 pKa = 3.19EE376 pKa = 4.36VEE378 pKa = 4.43RR379 pKa = 11.84EE380 pKa = 3.76FGEE383 pKa = 4.38YY384 pKa = 10.08EE385 pKa = 4.03NAADD389 pKa = 4.63PLDD392 pKa = 4.65EE393 pKa = 5.81IISEE397 pKa = 4.56LDD399 pKa = 3.29DD400 pKa = 4.02ASKK403 pKa = 10.99RR404 pKa = 11.84KK405 pKa = 9.51ILAKK409 pKa = 10.6LMNSTEE415 pKa = 4.18KK416 pKa = 10.69VVVMGQAPVTATMEE430 pKa = 4.18WEE432 pKa = 4.64KK433 pKa = 10.73EE434 pKa = 4.11NPMHH438 pKa = 7.41VLMEE442 pKa = 4.28NLVATVNEE450 pKa = 3.88QEE452 pKa = 4.41TIIRR456 pKa = 11.84RR457 pKa = 11.84HH458 pKa = 4.62GAEE461 pKa = 3.89INGLKK466 pKa = 10.63AKK468 pKa = 9.8VANLEE473 pKa = 4.08KK474 pKa = 10.76KK475 pKa = 8.96RR476 pKa = 11.84TFAVNEE482 pKa = 4.11SGLRR486 pKa = 11.84DD487 pKa = 3.87CLKK490 pKa = 10.73SAGDD494 pKa = 3.9KK495 pKa = 10.63LVSAANSIGNAIVKK509 pKa = 9.57VAEE512 pKa = 4.13EE513 pKa = 3.99VSPVRR518 pKa = 11.84EE519 pKa = 4.2GEE521 pKa = 3.74KK522 pKa = 10.92DD523 pKa = 3.49EE524 pKa = 6.39IEE526 pKa = 4.12MQEE529 pKa = 3.76ITDD532 pKa = 3.84SVEE535 pKa = 3.74TAEE538 pKa = 4.28EE539 pKa = 4.05VGKK542 pKa = 10.1NLEE545 pKa = 4.52PDD547 pKa = 3.91DD548 pKa = 4.01SQKK551 pKa = 10.96VYY553 pKa = 11.27NPTTMKK559 pKa = 10.08IKK561 pKa = 10.39CFHH564 pKa = 6.84CEE566 pKa = 3.33RR567 pKa = 11.84TFRR570 pKa = 11.84TMLGLKK576 pKa = 4.86THH578 pKa = 6.37KK579 pKa = 9.47MVKK582 pKa = 9.84HH583 pKa = 5.89EE584 pKa = 4.59SAFKK588 pKa = 10.62SDD590 pKa = 3.19FRR592 pKa = 11.84NPVKK596 pKa = 10.3MQGNAFLGKK605 pKa = 10.06RR606 pKa = 11.84YY607 pKa = 9.26PSRR610 pKa = 11.84NLSRR614 pKa = 11.84TSRR617 pKa = 11.84SSNDD621 pKa = 3.08SNQSMSRR628 pKa = 11.84SANPRR633 pKa = 11.84GAPYY637 pKa = 10.66TMEE640 pKa = 3.91QLKK643 pKa = 10.46KK644 pKa = 10.54VLSEE648 pKa = 4.46LLPTMVGQSSEE659 pKa = 4.08KK660 pKa = 9.4QQNN663 pKa = 3.0
MM1 pKa = 7.65EE2 pKa = 6.03FLRR5 pKa = 11.84QLTWFKK11 pKa = 10.24MAWLIAKK18 pKa = 9.55FYY20 pKa = 9.31MGFVMIMNISTILTFLVTLLVSLTWKK46 pKa = 9.39MIKK49 pKa = 9.94KK50 pKa = 9.57VALVDD55 pKa = 3.77LFYY58 pKa = 11.04EE59 pKa = 4.76GFGSVVDD66 pKa = 3.98GEE68 pKa = 4.78VIHH71 pKa = 6.03TVVEE75 pKa = 4.51AVRR78 pKa = 11.84EE79 pKa = 4.1AHH81 pKa = 6.19EE82 pKa = 4.07ATRR85 pKa = 11.84PVEE88 pKa = 4.1PPSWWEE94 pKa = 3.52EE95 pKa = 4.05AYY97 pKa = 10.66VWLMDD102 pKa = 3.52MAMDD106 pKa = 3.92NFDD109 pKa = 3.72VLRR112 pKa = 11.84ICLVMVAGLLVLLAVKK128 pKa = 10.33RR129 pKa = 11.84LVWRR133 pKa = 11.84RR134 pKa = 11.84AKK136 pKa = 10.53RR137 pKa = 11.84LVYY140 pKa = 9.48TMKK143 pKa = 10.65GFHH146 pKa = 6.86CEE148 pKa = 3.73AMVEE152 pKa = 4.29GSEE155 pKa = 4.45FGSGDD160 pKa = 3.16IPSFQIPMYY169 pKa = 9.8RR170 pKa = 11.84AGIFVSTFLGYY181 pKa = 9.87GVRR184 pKa = 11.84VDD186 pKa = 3.58NTLCVPTHH194 pKa = 5.4VVNLARR200 pKa = 11.84GGQLMADD207 pKa = 3.52TWILPRR213 pKa = 11.84TGILSKK219 pKa = 10.34LHH221 pKa = 6.95PDD223 pKa = 2.83VTYY226 pKa = 10.84YY227 pKa = 10.62YY228 pKa = 10.41LPEE231 pKa = 4.3KK232 pKa = 8.69TWSVLATSKK241 pKa = 10.78PKK243 pKa = 10.47VGPVMAAVVVSCAGPDD259 pKa = 4.01GVSHH263 pKa = 5.93GHH265 pKa = 6.78LRR267 pKa = 11.84QTPDD271 pKa = 3.38LGQVTYY277 pKa = 10.71SGSTLPGFSGAAYY290 pKa = 10.41FNGDD294 pKa = 3.89VLFGMHH300 pKa = 6.49TGVACEE306 pKa = 3.99RR307 pKa = 11.84NIGVAAHH314 pKa = 7.15LLQFEE319 pKa = 4.16ARR321 pKa = 11.84EE322 pKa = 4.1AFGQMEE328 pKa = 4.75YY329 pKa = 10.59KK330 pKa = 10.76GEE332 pKa = 4.15SPLNDD337 pKa = 3.32KK338 pKa = 8.61THH340 pKa = 7.12DD341 pKa = 3.9KK342 pKa = 10.83VLKK345 pKa = 7.08TQNRR349 pKa = 11.84VVTWKK354 pKa = 10.82SSDD357 pKa = 3.22ILEE360 pKa = 4.46KK361 pKa = 10.71AKK363 pKa = 10.76KK364 pKa = 9.4LWGPAGSSWADD375 pKa = 3.19EE376 pKa = 4.36VEE378 pKa = 4.43RR379 pKa = 11.84EE380 pKa = 3.76FGEE383 pKa = 4.38YY384 pKa = 10.08EE385 pKa = 4.03NAADD389 pKa = 4.63PLDD392 pKa = 4.65EE393 pKa = 5.81IISEE397 pKa = 4.56LDD399 pKa = 3.29DD400 pKa = 4.02ASKK403 pKa = 10.99RR404 pKa = 11.84KK405 pKa = 9.51ILAKK409 pKa = 10.6LMNSTEE415 pKa = 4.18KK416 pKa = 10.69VVVMGQAPVTATMEE430 pKa = 4.18WEE432 pKa = 4.64KK433 pKa = 10.73EE434 pKa = 4.11NPMHH438 pKa = 7.41VLMEE442 pKa = 4.28NLVATVNEE450 pKa = 3.88QEE452 pKa = 4.41TIIRR456 pKa = 11.84RR457 pKa = 11.84HH458 pKa = 4.62GAEE461 pKa = 3.89INGLKK466 pKa = 10.63AKK468 pKa = 9.8VANLEE473 pKa = 4.08KK474 pKa = 10.76KK475 pKa = 8.96RR476 pKa = 11.84TFAVNEE482 pKa = 4.11SGLRR486 pKa = 11.84DD487 pKa = 3.87CLKK490 pKa = 10.73SAGDD494 pKa = 3.9KK495 pKa = 10.63LVSAANSIGNAIVKK509 pKa = 9.57VAEE512 pKa = 4.13EE513 pKa = 3.99VSPVRR518 pKa = 11.84EE519 pKa = 4.2GEE521 pKa = 3.74KK522 pKa = 10.92DD523 pKa = 3.49EE524 pKa = 6.39IEE526 pKa = 4.12MQEE529 pKa = 3.76ITDD532 pKa = 3.84SVEE535 pKa = 3.74TAEE538 pKa = 4.28EE539 pKa = 4.05VGKK542 pKa = 10.1NLEE545 pKa = 4.52PDD547 pKa = 3.91DD548 pKa = 4.01SQKK551 pKa = 10.96VYY553 pKa = 11.27NPTTMKK559 pKa = 10.08IKK561 pKa = 10.39CFHH564 pKa = 6.84CEE566 pKa = 3.33RR567 pKa = 11.84TFRR570 pKa = 11.84TMLGLKK576 pKa = 4.86THH578 pKa = 6.37KK579 pKa = 9.47MVKK582 pKa = 9.84HH583 pKa = 5.89EE584 pKa = 4.59SAFKK588 pKa = 10.62SDD590 pKa = 3.19FRR592 pKa = 11.84NPVKK596 pKa = 10.3MQGNAFLGKK605 pKa = 10.06RR606 pKa = 11.84YY607 pKa = 9.26PSRR610 pKa = 11.84NLSRR614 pKa = 11.84TSRR617 pKa = 11.84SSNDD621 pKa = 3.08SNQSMSRR628 pKa = 11.84SANPRR633 pKa = 11.84GAPYY637 pKa = 10.66TMEE640 pKa = 3.91QLKK643 pKa = 10.46KK644 pKa = 10.54VLSEE648 pKa = 4.46LLPTMVGQSSEE659 pKa = 4.08KK660 pKa = 9.4QQNN663 pKa = 3.0
Molecular weight: 74.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1030 |
367 |
663 |
515.0 |
58.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.408 ± 1.048 | 1.553 ± 0.321 |
5.146 ± 0.853 | 7.184 ± 0.609 |
3.689 ± 0.075 | 6.311 ± 0.441 |
2.913 ± 0.601 | 4.466 ± 0.643 |
6.408 ± 0.49 | 8.932 ± 0.17 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.369 ± 0.144 | 3.495 ± 0.534 |
4.272 ± 0.184 | 2.913 ± 0.183 |
5.146 ± 0.295 | 6.505 ± 0.261 |
5.534 ± 0.322 | 9.417 ± 0.218 |
2.427 ± 0.431 | 2.913 ± 0.462 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
