
Pseudoplusia includens SNPV IE
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Baculoviridae; Alphabaculovirus; Chrysodeixis includens nucleopolyhedrovirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
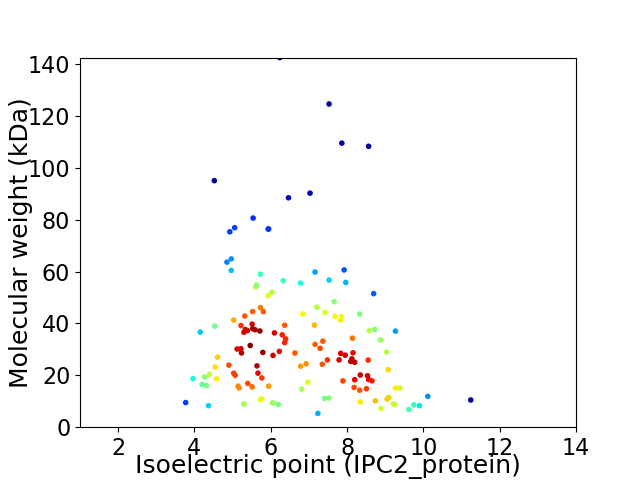
Virtual 2D-PAGE plot for 141 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
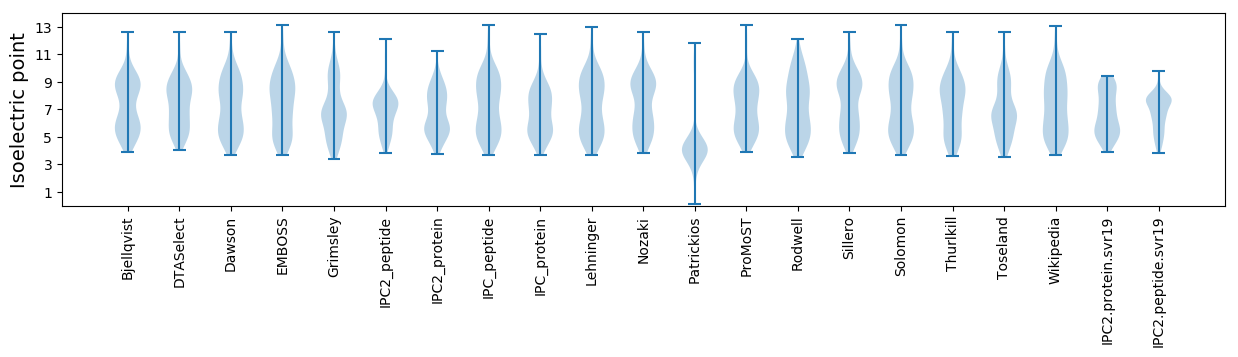
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4ZUP2|A0A0B4ZUP2_9ABAC Uncharacterized protein OS=Pseudoplusia includens SNPV IE OX=1592335 GN=ORF-111 PE=4 SV=1
MM1 pKa = 7.08FVSTEE6 pKa = 3.92EE7 pKa = 3.97VSRR10 pKa = 11.84GGLANIINMFAYY22 pKa = 9.65KK23 pKa = 10.41FIDD26 pKa = 3.26SDD28 pKa = 4.23YY29 pKa = 9.9EE30 pKa = 4.39NNVCTICEE38 pKa = 4.22SGFEE42 pKa = 4.0QYY44 pKa = 10.43EE45 pKa = 4.23PNVWTFRR52 pKa = 11.84VVEE55 pKa = 4.21KK56 pKa = 11.11NKK58 pKa = 10.35LNSIIYY64 pKa = 9.98CNICAEE70 pKa = 4.7EE71 pKa = 4.92IDD73 pKa = 5.99DD74 pKa = 5.7DD75 pKa = 6.54DD76 pKa = 4.7EE77 pKa = 6.33DD78 pKa = 4.3CFKK81 pKa = 10.79IALPASIEE89 pKa = 3.99DD90 pKa = 3.34QNRR93 pKa = 11.84FEE95 pKa = 4.43EE96 pKa = 4.37LLDD99 pKa = 3.8RR100 pKa = 11.84LIEE103 pKa = 4.5CGGVWLILNLVEE115 pKa = 5.7YY116 pKa = 10.31DD117 pKa = 3.22NCVNGNSIDD126 pKa = 3.81MEE128 pKa = 4.32TEE130 pKa = 3.55AFLKK134 pKa = 10.4VLRR137 pKa = 11.84FTSRR141 pKa = 11.84DD142 pKa = 3.03LDD144 pKa = 3.55RR145 pKa = 11.84PPNDD149 pKa = 3.3KK150 pKa = 10.42PGPINIIDD158 pKa = 4.12KK159 pKa = 10.49FCVSHH164 pKa = 6.57SLRR167 pKa = 11.84EE168 pKa = 3.96NYY170 pKa = 9.69INANILVDD178 pKa = 3.76KK179 pKa = 10.76LLNCTVDD186 pKa = 3.65MLPKK190 pKa = 10.38KK191 pKa = 10.55LNNCSRR197 pKa = 11.84EE198 pKa = 4.2KK199 pKa = 10.52FSHH202 pKa = 6.31ALSTFKK208 pKa = 10.82PNLLEE213 pKa = 5.62DD214 pKa = 3.76VLFSNTLVIPPEE226 pKa = 3.75NLYY229 pKa = 9.38TVMPTEE235 pKa = 4.93LNTLKK240 pKa = 10.55NQIYY244 pKa = 9.52EE245 pKa = 4.12VHH247 pKa = 5.99QLNDD251 pKa = 4.05DD252 pKa = 3.74EE253 pKa = 7.02DD254 pKa = 6.33DD255 pKa = 4.68NDD257 pKa = 4.0DD258 pKa = 3.74QEE260 pKa = 5.89KK261 pKa = 10.87DD262 pKa = 3.48FSSRR266 pKa = 11.84KK267 pKa = 8.21VSEE270 pKa = 3.88YY271 pKa = 10.98LIDD274 pKa = 3.87FLNDD278 pKa = 3.22FVSNYY283 pKa = 10.43NINPPNQHH291 pKa = 7.08AIMPYY296 pKa = 9.68CNSAAEE302 pKa = 3.93AMYY305 pKa = 11.02DD306 pKa = 3.34FDD308 pKa = 5.67LYY310 pKa = 11.53VYY312 pKa = 10.78APYY315 pKa = 11.04DD316 pKa = 3.36LL317 pKa = 5.52
MM1 pKa = 7.08FVSTEE6 pKa = 3.92EE7 pKa = 3.97VSRR10 pKa = 11.84GGLANIINMFAYY22 pKa = 9.65KK23 pKa = 10.41FIDD26 pKa = 3.26SDD28 pKa = 4.23YY29 pKa = 9.9EE30 pKa = 4.39NNVCTICEE38 pKa = 4.22SGFEE42 pKa = 4.0QYY44 pKa = 10.43EE45 pKa = 4.23PNVWTFRR52 pKa = 11.84VVEE55 pKa = 4.21KK56 pKa = 11.11NKK58 pKa = 10.35LNSIIYY64 pKa = 9.98CNICAEE70 pKa = 4.7EE71 pKa = 4.92IDD73 pKa = 5.99DD74 pKa = 5.7DD75 pKa = 6.54DD76 pKa = 4.7EE77 pKa = 6.33DD78 pKa = 4.3CFKK81 pKa = 10.79IALPASIEE89 pKa = 3.99DD90 pKa = 3.34QNRR93 pKa = 11.84FEE95 pKa = 4.43EE96 pKa = 4.37LLDD99 pKa = 3.8RR100 pKa = 11.84LIEE103 pKa = 4.5CGGVWLILNLVEE115 pKa = 5.7YY116 pKa = 10.31DD117 pKa = 3.22NCVNGNSIDD126 pKa = 3.81MEE128 pKa = 4.32TEE130 pKa = 3.55AFLKK134 pKa = 10.4VLRR137 pKa = 11.84FTSRR141 pKa = 11.84DD142 pKa = 3.03LDD144 pKa = 3.55RR145 pKa = 11.84PPNDD149 pKa = 3.3KK150 pKa = 10.42PGPINIIDD158 pKa = 4.12KK159 pKa = 10.49FCVSHH164 pKa = 6.57SLRR167 pKa = 11.84EE168 pKa = 3.96NYY170 pKa = 9.69INANILVDD178 pKa = 3.76KK179 pKa = 10.76LLNCTVDD186 pKa = 3.65MLPKK190 pKa = 10.38KK191 pKa = 10.55LNNCSRR197 pKa = 11.84EE198 pKa = 4.2KK199 pKa = 10.52FSHH202 pKa = 6.31ALSTFKK208 pKa = 10.82PNLLEE213 pKa = 5.62DD214 pKa = 3.76VLFSNTLVIPPEE226 pKa = 3.75NLYY229 pKa = 9.38TVMPTEE235 pKa = 4.93LNTLKK240 pKa = 10.55NQIYY244 pKa = 9.52EE245 pKa = 4.12VHH247 pKa = 5.99QLNDD251 pKa = 4.05DD252 pKa = 3.74EE253 pKa = 7.02DD254 pKa = 6.33DD255 pKa = 4.68NDD257 pKa = 4.0DD258 pKa = 3.74QEE260 pKa = 5.89KK261 pKa = 10.87DD262 pKa = 3.48FSSRR266 pKa = 11.84KK267 pKa = 8.21VSEE270 pKa = 3.88YY271 pKa = 10.98LIDD274 pKa = 3.87FLNDD278 pKa = 3.22FVSNYY283 pKa = 10.43NINPPNQHH291 pKa = 7.08AIMPYY296 pKa = 9.68CNSAAEE302 pKa = 3.93AMYY305 pKa = 11.02DD306 pKa = 3.34FDD308 pKa = 5.67LYY310 pKa = 11.53VYY312 pKa = 10.78APYY315 pKa = 11.04DD316 pKa = 3.36LL317 pKa = 5.52
Molecular weight: 36.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4ZUP8|A0A0B4ZUP8_9ABAC Alkaline exonuclease OS=Pseudoplusia includens SNPV IE OX=1592335 GN=ORF-121 PE=4 SV=1
MM1 pKa = 7.29VYY3 pKa = 10.2RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84SSGRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84SSGGQYY18 pKa = 9.83RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84SSGSRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84SSGGRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SSGGRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84SSGGQYY48 pKa = 9.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84SGSRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84SSGGRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84PGRR68 pKa = 11.84PRR70 pKa = 11.84GRR72 pKa = 11.84RR73 pKa = 11.84SSSSGSRR80 pKa = 11.84NPYY83 pKa = 7.93GYY85 pKa = 10.07SRR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84YY90 pKa = 8.97
MM1 pKa = 7.29VYY3 pKa = 10.2RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84SSGRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84SSGGQYY18 pKa = 9.83RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84SSGSRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84SSGGRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SSGGRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84SSGGQYY48 pKa = 9.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84SGSRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84SSGGRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84PGRR68 pKa = 11.84PRR70 pKa = 11.84GRR72 pKa = 11.84RR73 pKa = 11.84SSSSGSRR80 pKa = 11.84NPYY83 pKa = 7.93GYY85 pKa = 10.07SRR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84YY90 pKa = 8.97
Molecular weight: 10.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
41865 |
49 |
1209 |
296.9 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.615 ± 0.157 | 2.069 ± 0.126 |
6.628 ± 0.173 | 5.587 ± 0.14 |
4.878 ± 0.146 | 3.363 ± 0.145 |
2.186 ± 0.103 | 7.835 ± 0.136 |
6.908 ± 0.25 | 8.969 ± 0.189 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.654 ± 0.081 | 7.949 ± 0.197 |
3.449 ± 0.148 | 3.669 ± 0.155 |
5.054 ± 0.166 | 7.123 ± 0.217 |
5.453 ± 0.128 | 6.012 ± 0.148 |
0.731 ± 0.053 | 4.844 ± 0.151 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
