
Streptococcus satellite phage Javan286
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
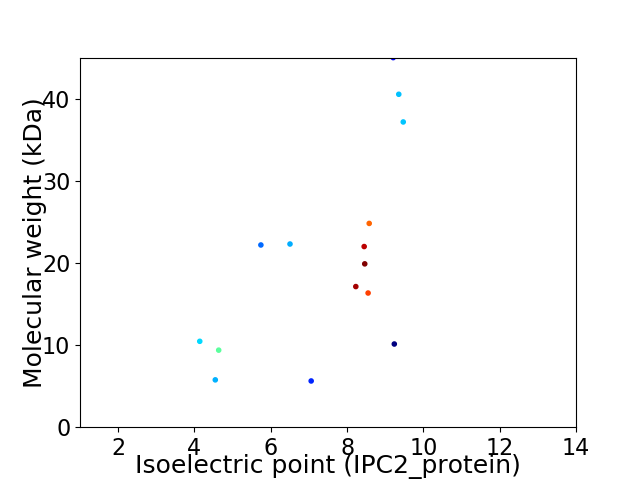
Virtual 2D-PAGE plot for 15 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
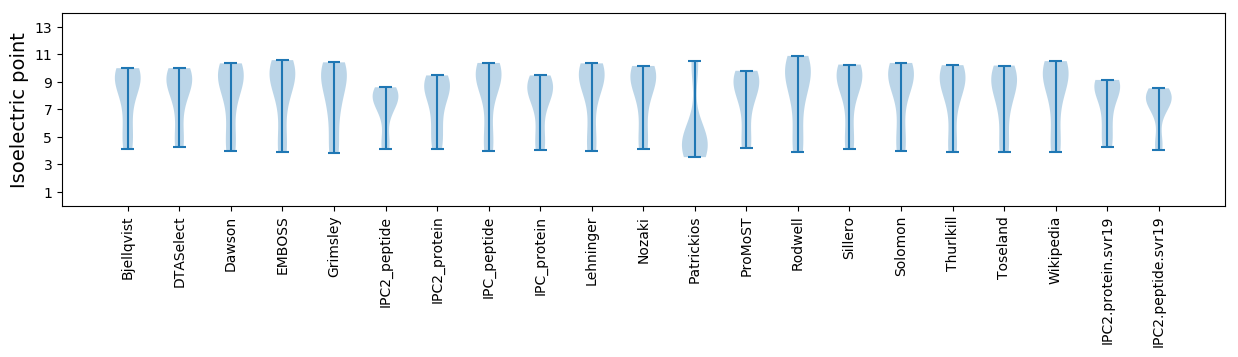
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZNC7|A0A4D5ZNC7_9VIRU Bro-N domain-containing protein OS=Streptococcus satellite phage Javan286 OX=2558608 GN=JavanS286_0003 PE=4 SV=1
MM1 pKa = 7.64LYY3 pKa = 9.76IEE5 pKa = 4.24QLEE8 pKa = 4.2EE9 pKa = 4.19EE10 pKa = 4.48IKK12 pKa = 10.9EE13 pKa = 4.06EE14 pKa = 3.98QQNSDD19 pKa = 3.7KK20 pKa = 10.9QPKK23 pKa = 8.11TMNEE27 pKa = 4.16LYY29 pKa = 10.56NLPGKK34 pKa = 9.9QEE36 pKa = 4.07GFDD39 pKa = 4.04GIAFLVFNCLGSFLMMLEE57 pKa = 4.6DD58 pKa = 6.32DD59 pKa = 4.91EE60 pKa = 5.77LHH62 pKa = 6.35CQPKK66 pKa = 10.06PIEE69 pKa = 4.38EE70 pKa = 4.85DD71 pKa = 3.12SDD73 pKa = 4.3LDD75 pKa = 3.72NEE77 pKa = 4.42ANKK80 pKa = 9.15TLAYY84 pKa = 10.5DD85 pKa = 3.95EE86 pKa = 4.69LVKK89 pKa = 10.78KK90 pKa = 10.67
MM1 pKa = 7.64LYY3 pKa = 9.76IEE5 pKa = 4.24QLEE8 pKa = 4.2EE9 pKa = 4.19EE10 pKa = 4.48IKK12 pKa = 10.9EE13 pKa = 4.06EE14 pKa = 3.98QQNSDD19 pKa = 3.7KK20 pKa = 10.9QPKK23 pKa = 8.11TMNEE27 pKa = 4.16LYY29 pKa = 10.56NLPGKK34 pKa = 9.9QEE36 pKa = 4.07GFDD39 pKa = 4.04GIAFLVFNCLGSFLMMLEE57 pKa = 4.6DD58 pKa = 6.32DD59 pKa = 4.91EE60 pKa = 5.77LHH62 pKa = 6.35CQPKK66 pKa = 10.06PIEE69 pKa = 4.38EE70 pKa = 4.85DD71 pKa = 3.12SDD73 pKa = 4.3LDD75 pKa = 3.72NEE77 pKa = 4.42ANKK80 pKa = 9.15TLAYY84 pKa = 10.5DD85 pKa = 3.95EE86 pKa = 4.69LVKK89 pKa = 10.78KK90 pKa = 10.67
Molecular weight: 10.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZI12|A0A4D5ZI12_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan286 OX=2558608 GN=JavanS286_0008 PE=4 SV=1
MM1 pKa = 7.05QNYY4 pKa = 6.87YY5 pKa = 8.99TPKK8 pKa = 10.23SKK10 pKa = 10.53HH11 pKa = 4.74LTLTEE16 pKa = 3.38RR17 pKa = 11.84RR18 pKa = 11.84MIEE21 pKa = 3.44RR22 pKa = 11.84WLQEE26 pKa = 3.71GLSNRR31 pKa = 11.84EE32 pKa = 3.29IARR35 pKa = 11.84RR36 pKa = 11.84LAKK39 pKa = 10.4APQTIHH45 pKa = 6.92NEE47 pKa = 4.04VKK49 pKa = 10.29RR50 pKa = 11.84GQVRR54 pKa = 11.84QQVRR58 pKa = 11.84KK59 pKa = 9.96GKK61 pKa = 10.81FEE63 pKa = 4.47VIYY66 pKa = 10.92SADD69 pKa = 3.74FAQKK73 pKa = 10.51AYY75 pKa = 10.43QNNRR79 pKa = 11.84KK80 pKa = 9.53RR81 pKa = 11.84SVKK84 pKa = 9.81QASLTKK90 pKa = 10.17GLKK93 pKa = 9.87EE94 pKa = 4.48KK95 pKa = 8.91ITHH98 pKa = 6.22YY99 pKa = 10.4IEE101 pKa = 4.27QKK103 pKa = 10.13YY104 pKa = 9.26SPEE107 pKa = 3.8MMVKK111 pKa = 10.16SKK113 pKa = 10.84GIPVPISTIYY123 pKa = 9.16YY124 pKa = 7.43WIHH127 pKa = 6.45HH128 pKa = 5.8GHH130 pKa = 6.98LGWTKK135 pKa = 11.34ADD137 pKa = 2.94ILYY140 pKa = 9.32PRR142 pKa = 11.84QEE144 pKa = 4.01KK145 pKa = 10.4AKK147 pKa = 10.11KK148 pKa = 9.6KK149 pKa = 9.53HH150 pKa = 5.94ASPNFKK156 pKa = 9.98PAGKK160 pKa = 9.89SIEE163 pKa = 4.1EE164 pKa = 4.07RR165 pKa = 11.84PEE167 pKa = 4.35SINKK171 pKa = 9.57RR172 pKa = 11.84EE173 pKa = 4.05NIGDD177 pKa = 3.79FEE179 pKa = 6.17IDD181 pKa = 3.31TVIQTRR187 pKa = 11.84AKK189 pKa = 10.38NEE191 pKa = 4.05CLLTLTDD198 pKa = 3.41RR199 pKa = 11.84KK200 pKa = 10.41SRR202 pKa = 11.84YY203 pKa = 9.43QIIRR207 pKa = 11.84LIPDD211 pKa = 3.59KK212 pKa = 11.17SAFSVNQALKK222 pKa = 10.72AILKK226 pKa = 9.3DD227 pKa = 3.54YY228 pKa = 11.14QMNSITADD236 pKa = 3.12NGVEE240 pKa = 4.25FSRR243 pKa = 11.84LAEE246 pKa = 4.26VFNPTHH252 pKa = 6.94IYY254 pKa = 9.03YY255 pKa = 10.46AHH257 pKa = 7.67PYY259 pKa = 9.47SSWEE263 pKa = 3.69RR264 pKa = 11.84GTNEE268 pKa = 3.29NHH270 pKa = 5.89NRR272 pKa = 11.84LIRR275 pKa = 11.84RR276 pKa = 11.84WLPKK280 pKa = 9.64GSKK283 pKa = 9.91NATQQQVAFIEE294 pKa = 4.16NWINNYY300 pKa = 8.73PKK302 pKa = 10.82KK303 pKa = 10.49LFNYY307 pKa = 9.38KK308 pKa = 9.99SPKK311 pKa = 10.06EE312 pKa = 3.87FLQAGG317 pKa = 3.78
MM1 pKa = 7.05QNYY4 pKa = 6.87YY5 pKa = 8.99TPKK8 pKa = 10.23SKK10 pKa = 10.53HH11 pKa = 4.74LTLTEE16 pKa = 3.38RR17 pKa = 11.84RR18 pKa = 11.84MIEE21 pKa = 3.44RR22 pKa = 11.84WLQEE26 pKa = 3.71GLSNRR31 pKa = 11.84EE32 pKa = 3.29IARR35 pKa = 11.84RR36 pKa = 11.84LAKK39 pKa = 10.4APQTIHH45 pKa = 6.92NEE47 pKa = 4.04VKK49 pKa = 10.29RR50 pKa = 11.84GQVRR54 pKa = 11.84QQVRR58 pKa = 11.84KK59 pKa = 9.96GKK61 pKa = 10.81FEE63 pKa = 4.47VIYY66 pKa = 10.92SADD69 pKa = 3.74FAQKK73 pKa = 10.51AYY75 pKa = 10.43QNNRR79 pKa = 11.84KK80 pKa = 9.53RR81 pKa = 11.84SVKK84 pKa = 9.81QASLTKK90 pKa = 10.17GLKK93 pKa = 9.87EE94 pKa = 4.48KK95 pKa = 8.91ITHH98 pKa = 6.22YY99 pKa = 10.4IEE101 pKa = 4.27QKK103 pKa = 10.13YY104 pKa = 9.26SPEE107 pKa = 3.8MMVKK111 pKa = 10.16SKK113 pKa = 10.84GIPVPISTIYY123 pKa = 9.16YY124 pKa = 7.43WIHH127 pKa = 6.45HH128 pKa = 5.8GHH130 pKa = 6.98LGWTKK135 pKa = 11.34ADD137 pKa = 2.94ILYY140 pKa = 9.32PRR142 pKa = 11.84QEE144 pKa = 4.01KK145 pKa = 10.4AKK147 pKa = 10.11KK148 pKa = 9.6KK149 pKa = 9.53HH150 pKa = 5.94ASPNFKK156 pKa = 9.98PAGKK160 pKa = 9.89SIEE163 pKa = 4.1EE164 pKa = 4.07RR165 pKa = 11.84PEE167 pKa = 4.35SINKK171 pKa = 9.57RR172 pKa = 11.84EE173 pKa = 4.05NIGDD177 pKa = 3.79FEE179 pKa = 6.17IDD181 pKa = 3.31TVIQTRR187 pKa = 11.84AKK189 pKa = 10.38NEE191 pKa = 4.05CLLTLTDD198 pKa = 3.41RR199 pKa = 11.84KK200 pKa = 10.41SRR202 pKa = 11.84YY203 pKa = 9.43QIIRR207 pKa = 11.84LIPDD211 pKa = 3.59KK212 pKa = 11.17SAFSVNQALKK222 pKa = 10.72AILKK226 pKa = 9.3DD227 pKa = 3.54YY228 pKa = 11.14QMNSITADD236 pKa = 3.12NGVEE240 pKa = 4.25FSRR243 pKa = 11.84LAEE246 pKa = 4.26VFNPTHH252 pKa = 6.94IYY254 pKa = 9.03YY255 pKa = 10.46AHH257 pKa = 7.67PYY259 pKa = 9.47SSWEE263 pKa = 3.69RR264 pKa = 11.84GTNEE268 pKa = 3.29NHH270 pKa = 5.89NRR272 pKa = 11.84LIRR275 pKa = 11.84RR276 pKa = 11.84WLPKK280 pKa = 9.64GSKK283 pKa = 9.91NATQQQVAFIEE294 pKa = 4.16NWINNYY300 pKa = 8.73PKK302 pKa = 10.82KK303 pKa = 10.49LFNYY307 pKa = 9.38KK308 pKa = 9.99SPKK311 pKa = 10.06EE312 pKa = 3.87FLQAGG317 pKa = 3.78
Molecular weight: 37.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2658 |
46 |
389 |
177.2 |
20.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.966 ± 0.403 | 1.129 ± 0.333 |
3.913 ± 0.579 | 7.111 ± 0.997 |
4.74 ± 0.755 | 4.552 ± 0.564 |
2.972 ± 0.417 | 7.788 ± 0.674 |
9.255 ± 0.724 | 11.249 ± 0.933 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.521 ± 0.34 | 5.944 ± 0.371 |
3.01 ± 0.353 | 4.176 ± 0.381 |
4.853 ± 0.565 | 5.982 ± 0.436 |
5.38 ± 0.58 | 4.816 ± 0.351 |
0.941 ± 0.154 | 4.703 ± 0.249 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
