
Sweet potato chlorotic fleck virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Quinvirinae; Carlavirus
Average proteome isoelectric point is 7.61
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
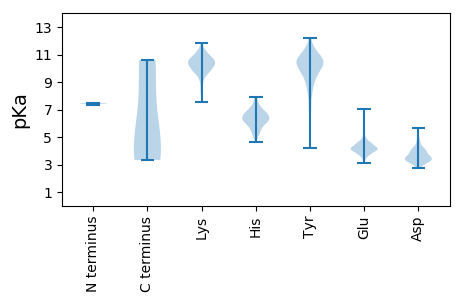
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q5L4G3|Q5L4G3_9VIRU 7 kDa protein OS=Sweet potato chlorotic fleck virus OX=263004 PE=3 SV=1
MM1 pKa = 7.35AAKK4 pKa = 10.0EE5 pKa = 4.1ADD7 pKa = 3.31TMEE10 pKa = 4.51KK11 pKa = 10.46VEE13 pKa = 4.14SSKK16 pKa = 11.15GKK18 pKa = 10.1EE19 pKa = 3.8KK20 pKa = 11.01AGGQTTTLEE29 pKa = 4.19GLNEE33 pKa = 3.89VKK35 pKa = 10.43AAQGEE40 pKa = 4.34ISDD43 pKa = 3.96QQLMKK48 pKa = 10.44HH49 pKa = 6.31DD50 pKa = 5.44ALIKK54 pKa = 9.69WYY56 pKa = 9.06MEE58 pKa = 3.83NYY60 pKa = 9.7RR61 pKa = 11.84PSDD64 pKa = 3.45VVNPLMQSGDD74 pKa = 3.37KK75 pKa = 10.77HH76 pKa = 5.13VTLSDD81 pKa = 3.53NLKK84 pKa = 10.07EE85 pKa = 4.1DD86 pKa = 3.15AANIYY91 pKa = 10.22SRR93 pKa = 11.84PNFNTLLKK101 pKa = 9.8WNRR104 pKa = 11.84EE105 pKa = 3.97PVSQSIATAEE115 pKa = 4.72DD116 pKa = 3.24IAQIEE121 pKa = 4.18AMLVGLGIPQEE132 pKa = 4.18RR133 pKa = 11.84VRR135 pKa = 11.84TAILDD140 pKa = 3.52IVLYY144 pKa = 9.83AAHH147 pKa = 6.83SSSSPQQLYY156 pKa = 10.59EE157 pKa = 3.93GDD159 pKa = 3.24IDD161 pKa = 5.02FRR163 pKa = 11.84DD164 pKa = 3.78ANFKK168 pKa = 10.13EE169 pKa = 4.69AISRR173 pKa = 11.84SSVAAVIKK181 pKa = 10.56DD182 pKa = 3.05KK183 pKa = 10.84STIRR187 pKa = 11.84KK188 pKa = 7.54VCRR191 pKa = 11.84LFAPVVWSYY200 pKa = 10.57MIINNEE206 pKa = 4.23PPSGWQAKK214 pKa = 8.58GFPEE218 pKa = 4.02NAKK221 pKa = 10.07FAAFDD226 pKa = 3.46TFDD229 pKa = 3.4FVTNHH234 pKa = 6.19AAIKK238 pKa = 8.77PLEE241 pKa = 4.88GIARR245 pKa = 11.84PPNSVEE251 pKa = 3.98YY252 pKa = 10.33IAAQTSKK259 pKa = 11.01RR260 pKa = 11.84ILIDD264 pKa = 3.11KK265 pKa = 10.12ARR267 pKa = 11.84RR268 pKa = 11.84NEE270 pKa = 3.8KK271 pKa = 10.17LSNYY275 pKa = 6.89EE276 pKa = 3.73ASVTGGQQACEE287 pKa = 3.93IKK289 pKa = 10.03TEE291 pKa = 4.13LKK293 pKa = 11.21GNGKK297 pKa = 9.85CKK299 pKa = 10.55
MM1 pKa = 7.35AAKK4 pKa = 10.0EE5 pKa = 4.1ADD7 pKa = 3.31TMEE10 pKa = 4.51KK11 pKa = 10.46VEE13 pKa = 4.14SSKK16 pKa = 11.15GKK18 pKa = 10.1EE19 pKa = 3.8KK20 pKa = 11.01AGGQTTTLEE29 pKa = 4.19GLNEE33 pKa = 3.89VKK35 pKa = 10.43AAQGEE40 pKa = 4.34ISDD43 pKa = 3.96QQLMKK48 pKa = 10.44HH49 pKa = 6.31DD50 pKa = 5.44ALIKK54 pKa = 9.69WYY56 pKa = 9.06MEE58 pKa = 3.83NYY60 pKa = 9.7RR61 pKa = 11.84PSDD64 pKa = 3.45VVNPLMQSGDD74 pKa = 3.37KK75 pKa = 10.77HH76 pKa = 5.13VTLSDD81 pKa = 3.53NLKK84 pKa = 10.07EE85 pKa = 4.1DD86 pKa = 3.15AANIYY91 pKa = 10.22SRR93 pKa = 11.84PNFNTLLKK101 pKa = 9.8WNRR104 pKa = 11.84EE105 pKa = 3.97PVSQSIATAEE115 pKa = 4.72DD116 pKa = 3.24IAQIEE121 pKa = 4.18AMLVGLGIPQEE132 pKa = 4.18RR133 pKa = 11.84VRR135 pKa = 11.84TAILDD140 pKa = 3.52IVLYY144 pKa = 9.83AAHH147 pKa = 6.83SSSSPQQLYY156 pKa = 10.59EE157 pKa = 3.93GDD159 pKa = 3.24IDD161 pKa = 5.02FRR163 pKa = 11.84DD164 pKa = 3.78ANFKK168 pKa = 10.13EE169 pKa = 4.69AISRR173 pKa = 11.84SSVAAVIKK181 pKa = 10.56DD182 pKa = 3.05KK183 pKa = 10.84STIRR187 pKa = 11.84KK188 pKa = 7.54VCRR191 pKa = 11.84LFAPVVWSYY200 pKa = 10.57MIINNEE206 pKa = 4.23PPSGWQAKK214 pKa = 8.58GFPEE218 pKa = 4.02NAKK221 pKa = 10.07FAAFDD226 pKa = 3.46TFDD229 pKa = 3.4FVTNHH234 pKa = 6.19AAIKK238 pKa = 8.77PLEE241 pKa = 4.88GIARR245 pKa = 11.84PPNSVEE251 pKa = 3.98YY252 pKa = 10.33IAAQTSKK259 pKa = 11.01RR260 pKa = 11.84ILIDD264 pKa = 3.11KK265 pKa = 10.12ARR267 pKa = 11.84RR268 pKa = 11.84NEE270 pKa = 3.8KK271 pKa = 10.17LSNYY275 pKa = 6.89EE276 pKa = 3.73ASVTGGQQACEE287 pKa = 3.93IKK289 pKa = 10.03TEE291 pKa = 4.13LKK293 pKa = 11.21GNGKK297 pKa = 9.85CKK299 pKa = 10.55
Molecular weight: 33.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5L4G2|Q5L4G2_9VIRU Capsid protein OS=Sweet potato chlorotic fleck virus OX=263004 PE=3 SV=1
MM1 pKa = 7.42GSASEE6 pKa = 4.07RR7 pKa = 11.84LPNFVATFLYY17 pKa = 10.64GRR19 pKa = 11.84GLLCDD24 pKa = 4.04WDD26 pKa = 4.18CCLLISSIVSNYY38 pKa = 8.6NKK40 pKa = 10.46GMRR43 pKa = 11.84TYY45 pKa = 11.23KK46 pKa = 10.44FFACGQSKK54 pKa = 10.38SAIKK58 pKa = 10.17RR59 pKa = 11.84RR60 pKa = 11.84ARR62 pKa = 11.84RR63 pKa = 11.84MQICPRR69 pKa = 11.84CAKK72 pKa = 10.04YY73 pKa = 10.58EE74 pKa = 4.09CGKK77 pKa = 10.21KK78 pKa = 10.23CEE80 pKa = 4.38PNTYY84 pKa = 10.21SQLQVKK90 pKa = 9.3EE91 pKa = 4.06LVTVGVNRR99 pKa = 11.84YY100 pKa = 4.18TTEE103 pKa = 4.27NITKK107 pKa = 10.17KK108 pKa = 8.31GTNVRR113 pKa = 11.84RR114 pKa = 11.84HH115 pKa = 6.1CKK117 pKa = 10.33NEE119 pKa = 3.72LEE121 pKa = 4.4FIKK124 pKa = 11.1YY125 pKa = 9.49EE126 pKa = 4.07SLKK129 pKa = 10.76VKK131 pKa = 10.5PKK133 pKa = 10.6
MM1 pKa = 7.42GSASEE6 pKa = 4.07RR7 pKa = 11.84LPNFVATFLYY17 pKa = 10.64GRR19 pKa = 11.84GLLCDD24 pKa = 4.04WDD26 pKa = 4.18CCLLISSIVSNYY38 pKa = 8.6NKK40 pKa = 10.46GMRR43 pKa = 11.84TYY45 pKa = 11.23KK46 pKa = 10.44FFACGQSKK54 pKa = 10.38SAIKK58 pKa = 10.17RR59 pKa = 11.84RR60 pKa = 11.84ARR62 pKa = 11.84RR63 pKa = 11.84MQICPRR69 pKa = 11.84CAKK72 pKa = 10.04YY73 pKa = 10.58EE74 pKa = 4.09CGKK77 pKa = 10.21KK78 pKa = 10.23CEE80 pKa = 4.38PNTYY84 pKa = 10.21SQLQVKK90 pKa = 9.3EE91 pKa = 4.06LVTVGVNRR99 pKa = 11.84YY100 pKa = 4.18TTEE103 pKa = 4.27NITKK107 pKa = 10.17KK108 pKa = 8.31GTNVRR113 pKa = 11.84RR114 pKa = 11.84HH115 pKa = 6.1CKK117 pKa = 10.33NEE119 pKa = 3.72LEE121 pKa = 4.4FIKK124 pKa = 11.1YY125 pKa = 9.49EE126 pKa = 4.07SLKK129 pKa = 10.76VKK131 pKa = 10.5PKK133 pKa = 10.6
Molecular weight: 15.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2937 |
67 |
2090 |
489.5 |
55.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.958 ± 0.913 | 2.656 ± 0.807 |
5.175 ± 0.565 | 6.878 ± 1.056 |
5.856 ± 0.933 | 6.435 ± 0.362 |
1.736 ± 0.233 | 5.993 ± 0.675 |
7.116 ± 0.701 | 9.057 ± 0.575 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.805 ± 0.254 | 4.699 ± 0.569 |
3.541 ± 0.588 | 2.826 ± 0.353 |
5.958 ± 0.589 | 7.729 ± 0.479 |
5.312 ± 0.525 | 6.639 ± 0.817 |
0.851 ± 0.166 | 3.779 ± 0.35 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
