
Blastococcus sp. CT_GayMR20
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; unclassified Blastococcus
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
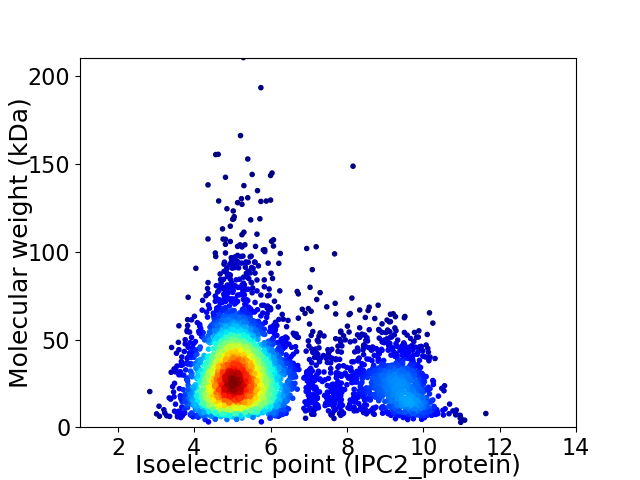
Virtual 2D-PAGE plot for 4483 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y9PS02|A0A4Y9PS02_9ACTN DegT/DnrJ/EryC1/StrS family aminotransferase OS=Blastococcus sp. CT_GayMR20 OX=2559609 GN=E4P40_14420 PE=3 SV=1
MM1 pKa = 8.19DD2 pKa = 5.53GPTEE6 pKa = 4.07PNQRR10 pKa = 11.84MVPSMRR16 pKa = 11.84SARR19 pKa = 11.84LTASALATAMLMFVAACGDD38 pKa = 3.88DD39 pKa = 3.93GGGGAADD46 pKa = 4.28GGNEE50 pKa = 4.1GGSGGGDD57 pKa = 3.39APRR60 pKa = 11.84IAAVFSGTTTDD71 pKa = 3.62ADD73 pKa = 3.94YY74 pKa = 10.73TFLGLEE80 pKa = 4.15ALQAAEE86 pKa = 4.14QEE88 pKa = 4.62HH89 pKa = 6.16GAEE92 pKa = 4.2TTYY95 pKa = 10.96SEE97 pKa = 4.66SVQVPDD103 pKa = 3.4AEE105 pKa = 4.23RR106 pKa = 11.84VLRR109 pKa = 11.84EE110 pKa = 4.0YY111 pKa = 10.97VADD114 pKa = 4.89GYY116 pKa = 8.55TVIWTHH122 pKa = 6.38GSQFYY127 pKa = 10.74DD128 pKa = 3.03ATVAVAEE135 pKa = 4.21EE136 pKa = 4.29APDD139 pKa = 3.49VVFIAEE145 pKa = 4.22QDD147 pKa = 3.68TEE149 pKa = 4.24PADD152 pKa = 3.56VPDD155 pKa = 4.26NVWVLDD161 pKa = 4.16RR162 pKa = 11.84NFHH165 pKa = 6.39LAFYY169 pKa = 9.81PLGVLAAAASQTGNIAYY186 pKa = 10.17LGGVTLPFSYY196 pKa = 10.81SEE198 pKa = 3.96VHH200 pKa = 6.63AVEE203 pKa = 4.05QALADD208 pKa = 4.02SGSDD212 pKa = 3.21ATFTPVWTGDD222 pKa = 3.43FNDD225 pKa = 3.91PTKK228 pKa = 10.65AQQFASQLLADD239 pKa = 4.61GNDD242 pKa = 3.41VVLTSLNLGAVGAFEE257 pKa = 4.62AVGDD261 pKa = 4.17AGPGVLMTAKK271 pKa = 9.5YY272 pKa = 8.6TDD274 pKa = 3.71KK275 pKa = 11.47SNLSADD281 pKa = 3.74HH282 pKa = 6.43YY283 pKa = 9.19LTSVLYY289 pKa = 10.32DD290 pKa = 3.66FSVMLDD296 pKa = 4.14DD297 pKa = 4.41ILTGIEE303 pKa = 4.02DD304 pKa = 4.47GEE306 pKa = 4.56TGGYY310 pKa = 10.57LGMSYY315 pKa = 10.34EE316 pKa = 4.29QGISLQTPQNVDD328 pKa = 2.98QAVIDD333 pKa = 3.9EE334 pKa = 4.46VEE336 pKa = 4.25QIVEE340 pKa = 3.83QLAAGEE346 pKa = 4.38IEE348 pKa = 4.32VEE350 pKa = 3.91RR351 pKa = 11.84DD352 pKa = 3.41VTPVEE357 pKa = 4.13
MM1 pKa = 8.19DD2 pKa = 5.53GPTEE6 pKa = 4.07PNQRR10 pKa = 11.84MVPSMRR16 pKa = 11.84SARR19 pKa = 11.84LTASALATAMLMFVAACGDD38 pKa = 3.88DD39 pKa = 3.93GGGGAADD46 pKa = 4.28GGNEE50 pKa = 4.1GGSGGGDD57 pKa = 3.39APRR60 pKa = 11.84IAAVFSGTTTDD71 pKa = 3.62ADD73 pKa = 3.94YY74 pKa = 10.73TFLGLEE80 pKa = 4.15ALQAAEE86 pKa = 4.14QEE88 pKa = 4.62HH89 pKa = 6.16GAEE92 pKa = 4.2TTYY95 pKa = 10.96SEE97 pKa = 4.66SVQVPDD103 pKa = 3.4AEE105 pKa = 4.23RR106 pKa = 11.84VLRR109 pKa = 11.84EE110 pKa = 4.0YY111 pKa = 10.97VADD114 pKa = 4.89GYY116 pKa = 8.55TVIWTHH122 pKa = 6.38GSQFYY127 pKa = 10.74DD128 pKa = 3.03ATVAVAEE135 pKa = 4.21EE136 pKa = 4.29APDD139 pKa = 3.49VVFIAEE145 pKa = 4.22QDD147 pKa = 3.68TEE149 pKa = 4.24PADD152 pKa = 3.56VPDD155 pKa = 4.26NVWVLDD161 pKa = 4.16RR162 pKa = 11.84NFHH165 pKa = 6.39LAFYY169 pKa = 9.81PLGVLAAAASQTGNIAYY186 pKa = 10.17LGGVTLPFSYY196 pKa = 10.81SEE198 pKa = 3.96VHH200 pKa = 6.63AVEE203 pKa = 4.05QALADD208 pKa = 4.02SGSDD212 pKa = 3.21ATFTPVWTGDD222 pKa = 3.43FNDD225 pKa = 3.91PTKK228 pKa = 10.65AQQFASQLLADD239 pKa = 4.61GNDD242 pKa = 3.41VVLTSLNLGAVGAFEE257 pKa = 4.62AVGDD261 pKa = 4.17AGPGVLMTAKK271 pKa = 9.5YY272 pKa = 8.6TDD274 pKa = 3.71KK275 pKa = 11.47SNLSADD281 pKa = 3.74HH282 pKa = 6.43YY283 pKa = 9.19LTSVLYY289 pKa = 10.32DD290 pKa = 3.66FSVMLDD296 pKa = 4.14DD297 pKa = 4.41ILTGIEE303 pKa = 4.02DD304 pKa = 4.47GEE306 pKa = 4.56TGGYY310 pKa = 10.57LGMSYY315 pKa = 10.34EE316 pKa = 4.29QGISLQTPQNVDD328 pKa = 2.98QAVIDD333 pKa = 3.9EE334 pKa = 4.46VEE336 pKa = 4.25QIVEE340 pKa = 3.83QLAAGEE346 pKa = 4.38IEE348 pKa = 4.32VEE350 pKa = 3.91RR351 pKa = 11.84DD352 pKa = 3.41VTPVEE357 pKa = 4.13
Molecular weight: 37.47 kDa
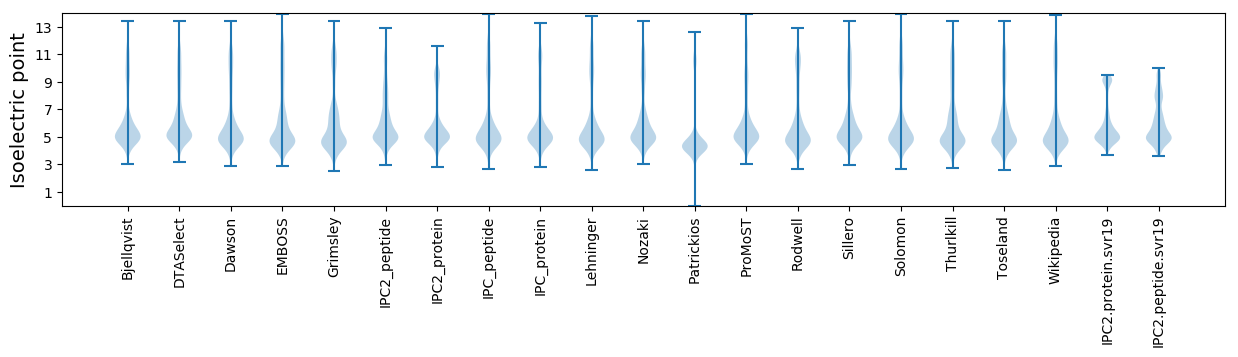
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9NPH4|A0A4Y9NPH4_9ACTN Uncharacterized protein OS=Blastococcus sp. CT_GayMR20 OX=2559609 GN=E4P40_22130 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1354267 |
24 |
1960 |
302.1 |
32.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.83 ± 0.058 | 0.678 ± 0.01 |
6.353 ± 0.03 | 5.724 ± 0.033 |
2.656 ± 0.023 | 9.513 ± 0.037 |
2.019 ± 0.018 | 3.115 ± 0.026 |
1.51 ± 0.023 | 10.674 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.699 ± 0.013 | 1.542 ± 0.017 |
6.12 ± 0.029 | 2.703 ± 0.019 |
8.084 ± 0.046 | 5.023 ± 0.029 |
5.907 ± 0.035 | 9.629 ± 0.039 |
1.452 ± 0.014 | 1.769 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
