
Serinicoccus chungangensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Ornithinimicrobiaceae; Serinicoccus
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
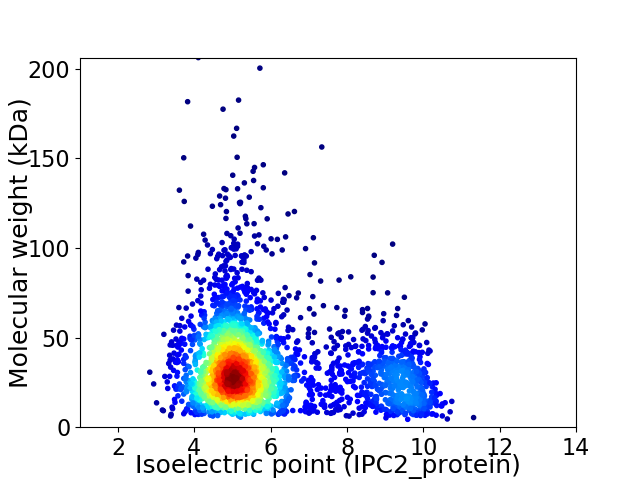
Virtual 2D-PAGE plot for 3100 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W8IHA6|A0A0W8IHA6_9MICO Uncharacterized protein OS=Serinicoccus chungangensis OX=767452 GN=AVL62_06395 PE=4 SV=1
MM1 pKa = 7.1QNVPRR6 pKa = 11.84RR7 pKa = 11.84AVAAASTAALAVTGLVALSPAVQAAGPGLVVNEE40 pKa = 4.28VYY42 pKa = 10.83GGGGNSGAVYY52 pKa = 8.72THH54 pKa = 7.26DD55 pKa = 3.54FVEE58 pKa = 4.92LVNSTDD64 pKa = 3.87APVDD68 pKa = 3.73LDD70 pKa = 4.18GYY72 pKa = 10.06AVSYY76 pKa = 10.23YY77 pKa = 10.61SSSGNLGNACPLSGTVEE94 pKa = 4.1PGASYY99 pKa = 10.32LIQQNPGSGGTTPLPDD115 pKa = 4.48PDD117 pKa = 4.19IEE119 pKa = 5.47CGASMSGSNGIVEE132 pKa = 4.71LSLDD136 pKa = 3.9GEE138 pKa = 4.76VVDD141 pKa = 4.71LVGYY145 pKa = 10.28GSATRR150 pKa = 11.84FEE152 pKa = 4.82GSGAAPGLSNTTSASRR168 pKa = 11.84SDD170 pKa = 5.22AVDD173 pKa = 3.38TDD175 pKa = 4.61DD176 pKa = 4.57NAADD180 pKa = 4.07FTRR183 pKa = 11.84GEE185 pKa = 4.14PTPTGSGGGTDD196 pKa = 4.74PEE198 pKa = 4.74PEE200 pKa = 4.68PDD202 pKa = 4.21PEE204 pKa = 4.77PVDD207 pKa = 3.44TTIAQIQGTGATSPLEE223 pKa = 4.09GQPVNTSGVVTAAYY237 pKa = 7.02PTGGFNGVYY246 pKa = 9.96LQTAGSGGAAKK257 pKa = 10.6APGDD261 pKa = 3.65ASDD264 pKa = 4.91GVFLYY269 pKa = 10.88SSWAADD275 pKa = 3.29NLEE278 pKa = 4.33IGDD281 pKa = 4.26CVEE284 pKa = 4.18VEE286 pKa = 4.1DD287 pKa = 5.57GEE289 pKa = 4.61VVEE292 pKa = 4.65YY293 pKa = 11.24NGLTEE298 pKa = 3.84ISGGFVTVVDD308 pKa = 3.95GCEE311 pKa = 3.78AVVPTVLSSLPATDD325 pKa = 3.86AEE327 pKa = 4.46KK328 pKa = 10.56EE329 pKa = 4.38VYY331 pKa = 10.09EE332 pKa = 4.37GMLVQPEE339 pKa = 4.26GTYY342 pKa = 10.34TITNNYY348 pKa = 8.78QLNQYY353 pKa = 8.5GQLGLAVGDD362 pKa = 3.85EE363 pKa = 4.48PLYY366 pKa = 10.59QATDD370 pKa = 3.41QVLPGPEE377 pKa = 3.7AEE379 pKa = 5.09AYY381 pKa = 9.34QAANQEE387 pKa = 4.37KK388 pKa = 10.68YY389 pKa = 8.76ITLDD393 pKa = 4.52DD394 pKa = 4.57GSSWDD399 pKa = 3.6YY400 pKa = 11.26VRR402 pKa = 11.84NSTAQDD408 pKa = 3.27SPLPYY413 pKa = 10.42LSAQEE418 pKa = 3.88PHH420 pKa = 6.01RR421 pKa = 11.84TGSQVTFEE429 pKa = 4.27QPVVLDD435 pKa = 5.5YY436 pKa = 11.09RR437 pKa = 11.84FQWNYY442 pKa = 9.26QPVGQVVGSDD452 pKa = 3.81SPVDD456 pKa = 4.3PISSEE461 pKa = 3.74NDD463 pKa = 2.98RR464 pKa = 11.84EE465 pKa = 4.18YY466 pKa = 11.18AAPEE470 pKa = 3.77VGGNIQIGAFNVLNYY485 pKa = 10.2FSDD488 pKa = 3.97LGQDD492 pKa = 3.83EE493 pKa = 4.64EE494 pKa = 5.12DD495 pKa = 3.9CDD497 pKa = 4.26FFADD501 pKa = 4.04RR502 pKa = 11.84FGNPVATDD510 pKa = 3.8FCEE513 pKa = 4.62VRR515 pKa = 11.84GAWSEE520 pKa = 3.95QAFADD525 pKa = 3.89QQAKK529 pKa = 10.07LVTAINGTDD538 pKa = 3.57AEE540 pKa = 4.52VLALMEE546 pKa = 4.69IEE548 pKa = 4.24NSAALSYY555 pKa = 10.96VDD557 pKa = 4.25HH558 pKa = 7.11PRR560 pKa = 11.84DD561 pKa = 3.19KK562 pKa = 11.32ALADD566 pKa = 3.79LVAALNAEE574 pKa = 4.25AGEE577 pKa = 4.17QRR579 pKa = 11.84WAYY582 pKa = 10.15APSPTVTPPNEE593 pKa = 4.14DD594 pKa = 3.45VIRR597 pKa = 11.84TAFIYY602 pKa = 10.84DD603 pKa = 3.65PDD605 pKa = 3.82QVQLLGPSMIQLDD618 pKa = 3.89DD619 pKa = 3.69AFANARR625 pKa = 11.84YY626 pKa = 8.83PLAQKK631 pKa = 10.36FKK633 pKa = 10.33ATGTGKK639 pKa = 9.81PFVAVANHH647 pKa = 6.25FKK649 pKa = 11.01SKK651 pKa = 10.83GSGEE655 pKa = 4.27DD656 pKa = 3.57DD657 pKa = 3.4GTGQGNANPSRR668 pKa = 11.84IAQAEE673 pKa = 4.06ALTAWSEE680 pKa = 4.21EE681 pKa = 3.98MFEE684 pKa = 4.69DD685 pKa = 4.41EE686 pKa = 6.5AIFLMGDD693 pKa = 3.48FNAYY697 pKa = 9.82SRR699 pKa = 11.84EE700 pKa = 3.97DD701 pKa = 3.45PVRR704 pKa = 11.84IIEE707 pKa = 4.39DD708 pKa = 3.03AGYY711 pKa = 8.96TNLARR716 pKa = 11.84ADD718 pKa = 4.22EE719 pKa = 4.91PDD721 pKa = 3.07SMTYY725 pKa = 10.04QFSGRR730 pKa = 11.84LGSLDD735 pKa = 3.49HH736 pKa = 6.92VFANEE741 pKa = 3.45KK742 pKa = 10.04AQRR745 pKa = 11.84LVTGAGVWDD754 pKa = 3.79INADD758 pKa = 3.44EE759 pKa = 5.23SIAFQYY765 pKa = 10.49SRR767 pKa = 11.84RR768 pKa = 11.84NYY770 pKa = 10.23NVVDD774 pKa = 4.23FHH776 pKa = 9.45SDD778 pKa = 3.36DD779 pKa = 3.58QFASSDD785 pKa = 3.68HH786 pKa = 7.02DD787 pKa = 3.92PVLVGLDD794 pKa = 3.06TGNRR798 pKa = 11.84GKK800 pKa = 10.93GKK802 pKa = 10.06NN803 pKa = 3.34
MM1 pKa = 7.1QNVPRR6 pKa = 11.84RR7 pKa = 11.84AVAAASTAALAVTGLVALSPAVQAAGPGLVVNEE40 pKa = 4.28VYY42 pKa = 10.83GGGGNSGAVYY52 pKa = 8.72THH54 pKa = 7.26DD55 pKa = 3.54FVEE58 pKa = 4.92LVNSTDD64 pKa = 3.87APVDD68 pKa = 3.73LDD70 pKa = 4.18GYY72 pKa = 10.06AVSYY76 pKa = 10.23YY77 pKa = 10.61SSSGNLGNACPLSGTVEE94 pKa = 4.1PGASYY99 pKa = 10.32LIQQNPGSGGTTPLPDD115 pKa = 4.48PDD117 pKa = 4.19IEE119 pKa = 5.47CGASMSGSNGIVEE132 pKa = 4.71LSLDD136 pKa = 3.9GEE138 pKa = 4.76VVDD141 pKa = 4.71LVGYY145 pKa = 10.28GSATRR150 pKa = 11.84FEE152 pKa = 4.82GSGAAPGLSNTTSASRR168 pKa = 11.84SDD170 pKa = 5.22AVDD173 pKa = 3.38TDD175 pKa = 4.61DD176 pKa = 4.57NAADD180 pKa = 4.07FTRR183 pKa = 11.84GEE185 pKa = 4.14PTPTGSGGGTDD196 pKa = 4.74PEE198 pKa = 4.74PEE200 pKa = 4.68PDD202 pKa = 4.21PEE204 pKa = 4.77PVDD207 pKa = 3.44TTIAQIQGTGATSPLEE223 pKa = 4.09GQPVNTSGVVTAAYY237 pKa = 7.02PTGGFNGVYY246 pKa = 9.96LQTAGSGGAAKK257 pKa = 10.6APGDD261 pKa = 3.65ASDD264 pKa = 4.91GVFLYY269 pKa = 10.88SSWAADD275 pKa = 3.29NLEE278 pKa = 4.33IGDD281 pKa = 4.26CVEE284 pKa = 4.18VEE286 pKa = 4.1DD287 pKa = 5.57GEE289 pKa = 4.61VVEE292 pKa = 4.65YY293 pKa = 11.24NGLTEE298 pKa = 3.84ISGGFVTVVDD308 pKa = 3.95GCEE311 pKa = 3.78AVVPTVLSSLPATDD325 pKa = 3.86AEE327 pKa = 4.46KK328 pKa = 10.56EE329 pKa = 4.38VYY331 pKa = 10.09EE332 pKa = 4.37GMLVQPEE339 pKa = 4.26GTYY342 pKa = 10.34TITNNYY348 pKa = 8.78QLNQYY353 pKa = 8.5GQLGLAVGDD362 pKa = 3.85EE363 pKa = 4.48PLYY366 pKa = 10.59QATDD370 pKa = 3.41QVLPGPEE377 pKa = 3.7AEE379 pKa = 5.09AYY381 pKa = 9.34QAANQEE387 pKa = 4.37KK388 pKa = 10.68YY389 pKa = 8.76ITLDD393 pKa = 4.52DD394 pKa = 4.57GSSWDD399 pKa = 3.6YY400 pKa = 11.26VRR402 pKa = 11.84NSTAQDD408 pKa = 3.27SPLPYY413 pKa = 10.42LSAQEE418 pKa = 3.88PHH420 pKa = 6.01RR421 pKa = 11.84TGSQVTFEE429 pKa = 4.27QPVVLDD435 pKa = 5.5YY436 pKa = 11.09RR437 pKa = 11.84FQWNYY442 pKa = 9.26QPVGQVVGSDD452 pKa = 3.81SPVDD456 pKa = 4.3PISSEE461 pKa = 3.74NDD463 pKa = 2.98RR464 pKa = 11.84EE465 pKa = 4.18YY466 pKa = 11.18AAPEE470 pKa = 3.77VGGNIQIGAFNVLNYY485 pKa = 10.2FSDD488 pKa = 3.97LGQDD492 pKa = 3.83EE493 pKa = 4.64EE494 pKa = 5.12DD495 pKa = 3.9CDD497 pKa = 4.26FFADD501 pKa = 4.04RR502 pKa = 11.84FGNPVATDD510 pKa = 3.8FCEE513 pKa = 4.62VRR515 pKa = 11.84GAWSEE520 pKa = 3.95QAFADD525 pKa = 3.89QQAKK529 pKa = 10.07LVTAINGTDD538 pKa = 3.57AEE540 pKa = 4.52VLALMEE546 pKa = 4.69IEE548 pKa = 4.24NSAALSYY555 pKa = 10.96VDD557 pKa = 4.25HH558 pKa = 7.11PRR560 pKa = 11.84DD561 pKa = 3.19KK562 pKa = 11.32ALADD566 pKa = 3.79LVAALNAEE574 pKa = 4.25AGEE577 pKa = 4.17QRR579 pKa = 11.84WAYY582 pKa = 10.15APSPTVTPPNEE593 pKa = 4.14DD594 pKa = 3.45VIRR597 pKa = 11.84TAFIYY602 pKa = 10.84DD603 pKa = 3.65PDD605 pKa = 3.82QVQLLGPSMIQLDD618 pKa = 3.89DD619 pKa = 3.69AFANARR625 pKa = 11.84YY626 pKa = 8.83PLAQKK631 pKa = 10.36FKK633 pKa = 10.33ATGTGKK639 pKa = 9.81PFVAVANHH647 pKa = 6.25FKK649 pKa = 11.01SKK651 pKa = 10.83GSGEE655 pKa = 4.27DD656 pKa = 3.57DD657 pKa = 3.4GTGQGNANPSRR668 pKa = 11.84IAQAEE673 pKa = 4.06ALTAWSEE680 pKa = 4.21EE681 pKa = 3.98MFEE684 pKa = 4.69DD685 pKa = 4.41EE686 pKa = 6.5AIFLMGDD693 pKa = 3.48FNAYY697 pKa = 9.82SRR699 pKa = 11.84EE700 pKa = 3.97DD701 pKa = 3.45PVRR704 pKa = 11.84IIEE707 pKa = 4.39DD708 pKa = 3.03AGYY711 pKa = 8.96TNLARR716 pKa = 11.84ADD718 pKa = 4.22EE719 pKa = 4.91PDD721 pKa = 3.07SMTYY725 pKa = 10.04QFSGRR730 pKa = 11.84LGSLDD735 pKa = 3.49HH736 pKa = 6.92VFANEE741 pKa = 3.45KK742 pKa = 10.04AQRR745 pKa = 11.84LVTGAGVWDD754 pKa = 3.79INADD758 pKa = 3.44EE759 pKa = 5.23SIAFQYY765 pKa = 10.49SRR767 pKa = 11.84RR768 pKa = 11.84NYY770 pKa = 10.23NVVDD774 pKa = 4.23FHH776 pKa = 9.45SDD778 pKa = 3.36DD779 pKa = 3.58QFASSDD785 pKa = 3.68HH786 pKa = 7.02DD787 pKa = 3.92PVLVGLDD794 pKa = 3.06TGNRR798 pKa = 11.84GKK800 pKa = 10.93GKK802 pKa = 10.06NN803 pKa = 3.34
Molecular weight: 84.58 kDa
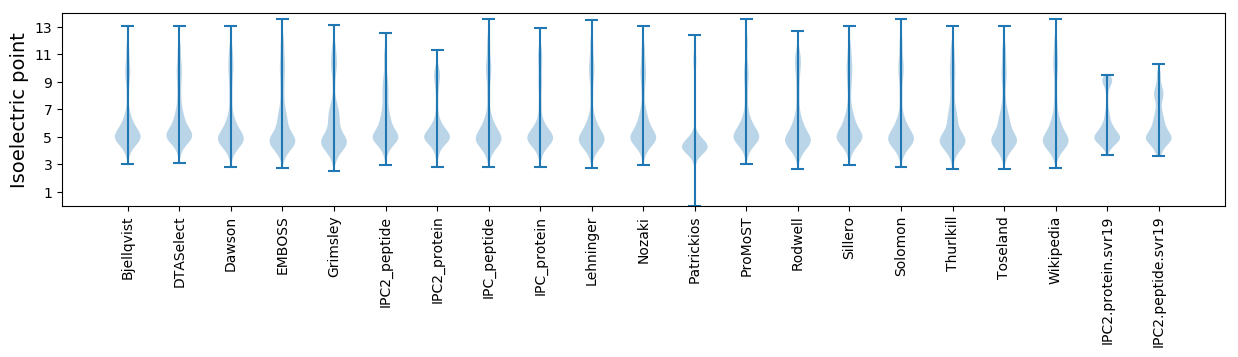
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W8I4D2|A0A0W8I4D2_9MICO Uncharacterized protein OS=Serinicoccus chungangensis OX=767452 GN=AVL62_14825 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILNARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.28GRR40 pKa = 11.84SSISAA45 pKa = 3.4
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILNARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.28GRR40 pKa = 11.84SSISAA45 pKa = 3.4
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1026549 |
37 |
1907 |
331.1 |
35.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.987 ± 0.061 | 0.655 ± 0.012 |
6.416 ± 0.038 | 6.192 ± 0.043 |
2.481 ± 0.027 | 9.712 ± 0.041 |
2.288 ± 0.026 | 2.871 ± 0.031 |
1.32 ± 0.028 | 10.63 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.84 ± 0.017 | 1.355 ± 0.02 |
5.88 ± 0.032 | 3.104 ± 0.023 |
8.053 ± 0.059 | 4.965 ± 0.031 |
6.069 ± 0.033 | 9.744 ± 0.047 |
1.627 ± 0.019 | 1.811 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
