
Wuhan flea virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
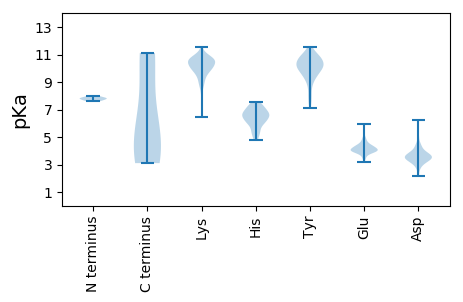
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0P0QKU2|A0A0P0QKU2_9VIRU Putative capsid protein OS=Wuhan flea virus OX=1746071 GN=VP2 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.62SNLCFIILTLLTTNILGKK20 pKa = 10.45DD21 pKa = 3.27RR22 pKa = 11.84DD23 pKa = 3.95CGKK26 pKa = 10.46FSEE29 pKa = 4.16NWRR32 pKa = 11.84ISACASGLKK41 pKa = 9.19QANLISDD48 pKa = 4.19ATYY51 pKa = 11.09VEE53 pKa = 4.49YY54 pKa = 10.81LLKK57 pKa = 10.8AIIATNGPNDD67 pKa = 3.95FYY69 pKa = 11.45GAGMTTGLDD78 pKa = 3.41TQLVIGKK85 pKa = 8.43MSTPVLSSGLSFNYY99 pKa = 9.68HH100 pKa = 6.63PSSMYY105 pKa = 10.09YY106 pKa = 9.74EE107 pKa = 4.01RR108 pKa = 11.84HH109 pKa = 5.62LVDD112 pKa = 5.09LGVKK116 pKa = 10.1FDD118 pKa = 4.06RR119 pKa = 11.84PVALFTIDD127 pKa = 3.22TMGSFPEE134 pKa = 4.15WTDD137 pKa = 3.09TMFMKK142 pKa = 9.87FVPTGSEE149 pKa = 3.19TRR151 pKa = 11.84YY152 pKa = 9.57AAFRR156 pKa = 11.84AGDD159 pKa = 3.85GSPVNIHH166 pKa = 5.49GQEE169 pKa = 3.74HH170 pKa = 6.48CADD173 pKa = 3.4FTKK176 pKa = 10.28WVDD179 pKa = 3.34SKK181 pKa = 11.55LPEE184 pKa = 4.53KK185 pKa = 10.66KK186 pKa = 9.89QGLAIHH192 pKa = 6.56MSQTLWCPVLKK203 pKa = 10.59DD204 pKa = 3.89GIMEE208 pKa = 4.1IVVAKK213 pKa = 10.58LNEE216 pKa = 3.96EE217 pKa = 3.96HH218 pKa = 7.28RR219 pKa = 11.84IAPNGEE225 pKa = 4.08SEE227 pKa = 4.33KK228 pKa = 10.89VLIVEE233 pKa = 4.24WAIWGSGAYY242 pKa = 10.15GDD244 pKa = 4.03VPEE247 pKa = 5.27EE248 pKa = 4.09YY249 pKa = 10.69DD250 pKa = 3.65SVFYY254 pKa = 11.12
MM1 pKa = 7.69KK2 pKa = 10.62SNLCFIILTLLTTNILGKK20 pKa = 10.45DD21 pKa = 3.27RR22 pKa = 11.84DD23 pKa = 3.95CGKK26 pKa = 10.46FSEE29 pKa = 4.16NWRR32 pKa = 11.84ISACASGLKK41 pKa = 9.19QANLISDD48 pKa = 4.19ATYY51 pKa = 11.09VEE53 pKa = 4.49YY54 pKa = 10.81LLKK57 pKa = 10.8AIIATNGPNDD67 pKa = 3.95FYY69 pKa = 11.45GAGMTTGLDD78 pKa = 3.41TQLVIGKK85 pKa = 8.43MSTPVLSSGLSFNYY99 pKa = 9.68HH100 pKa = 6.63PSSMYY105 pKa = 10.09YY106 pKa = 9.74EE107 pKa = 4.01RR108 pKa = 11.84HH109 pKa = 5.62LVDD112 pKa = 5.09LGVKK116 pKa = 10.1FDD118 pKa = 4.06RR119 pKa = 11.84PVALFTIDD127 pKa = 3.22TMGSFPEE134 pKa = 4.15WTDD137 pKa = 3.09TMFMKK142 pKa = 9.87FVPTGSEE149 pKa = 3.19TRR151 pKa = 11.84YY152 pKa = 9.57AAFRR156 pKa = 11.84AGDD159 pKa = 3.85GSPVNIHH166 pKa = 5.49GQEE169 pKa = 3.74HH170 pKa = 6.48CADD173 pKa = 3.4FTKK176 pKa = 10.28WVDD179 pKa = 3.34SKK181 pKa = 11.55LPEE184 pKa = 4.53KK185 pKa = 10.66KK186 pKa = 9.89QGLAIHH192 pKa = 6.56MSQTLWCPVLKK203 pKa = 10.59DD204 pKa = 3.89GIMEE208 pKa = 4.1IVVAKK213 pKa = 10.58LNEE216 pKa = 3.96EE217 pKa = 3.96HH218 pKa = 7.28RR219 pKa = 11.84IAPNGEE225 pKa = 4.08SEE227 pKa = 4.33KK228 pKa = 10.89VLIVEE233 pKa = 4.24WAIWGSGAYY242 pKa = 10.15GDD244 pKa = 4.03VPEE247 pKa = 5.27EE248 pKa = 4.09YY249 pKa = 10.69DD250 pKa = 3.65SVFYY254 pKa = 11.12
Molecular weight: 28.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0QKM5|A0A0P0QKM5_9VIRU Uncharacterized protein OS=Wuhan flea virus OX=1746071 GN=VP3 PE=4 SV=1
MM1 pKa = 7.99RR2 pKa = 11.84ICVCLILLIVSLTHH16 pKa = 7.36ASTSQQQARR25 pKa = 11.84SQEE28 pKa = 4.17LKK30 pKa = 10.87SEE32 pKa = 4.27LEE34 pKa = 4.32KK35 pKa = 10.73AWKK38 pKa = 10.13LDD40 pKa = 3.71FEE42 pKa = 4.8GLSLDD47 pKa = 3.47PTAWSSTLAGKK58 pKa = 10.01LSKK61 pKa = 10.73VAVGVGYY68 pKa = 11.34AMTLPMSLAHH78 pKa = 5.97VVGFGLFLFGLVRR91 pKa = 11.84LTWGIVGTSCLALYY105 pKa = 8.93GWKK108 pKa = 9.0YY109 pKa = 10.07HH110 pKa = 6.06KK111 pKa = 10.36RR112 pKa = 11.84SFLVAGVGLTYY123 pKa = 11.27ALAFGLSTLYY133 pKa = 10.99
MM1 pKa = 7.99RR2 pKa = 11.84ICVCLILLIVSLTHH16 pKa = 7.36ASTSQQQARR25 pKa = 11.84SQEE28 pKa = 4.17LKK30 pKa = 10.87SEE32 pKa = 4.27LEE34 pKa = 4.32KK35 pKa = 10.73AWKK38 pKa = 10.13LDD40 pKa = 3.71FEE42 pKa = 4.8GLSLDD47 pKa = 3.47PTAWSSTLAGKK58 pKa = 10.01LSKK61 pKa = 10.73VAVGVGYY68 pKa = 11.34AMTLPMSLAHH78 pKa = 5.97VVGFGLFLFGLVRR91 pKa = 11.84LTWGIVGTSCLALYY105 pKa = 8.93GWKK108 pKa = 9.0YY109 pKa = 10.07HH110 pKa = 6.06KK111 pKa = 10.36RR112 pKa = 11.84SFLVAGVGLTYY123 pKa = 11.27ALAFGLSTLYY133 pKa = 10.99
Molecular weight: 14.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2939 |
133 |
911 |
489.8 |
55.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.397 ± 0.619 | 1.395 ± 0.229 |
4.389 ± 0.416 | 6.635 ± 0.363 |
3.505 ± 0.253 | 8.098 ± 0.319 |
2.28 ± 0.213 | 6.703 ± 0.588 |
5.444 ± 0.735 | 9.969 ± 0.693 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.062 ± 0.216 | 3.607 ± 0.231 |
3.981 ± 0.332 | 2.79 ± 0.168 |
6.261 ± 0.736 | 5.648 ± 0.441 |
6.805 ± 0.476 | 6.975 ± 0.434 |
2.382 ± 0.389 | 3.675 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
