
Persimmon virus B
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
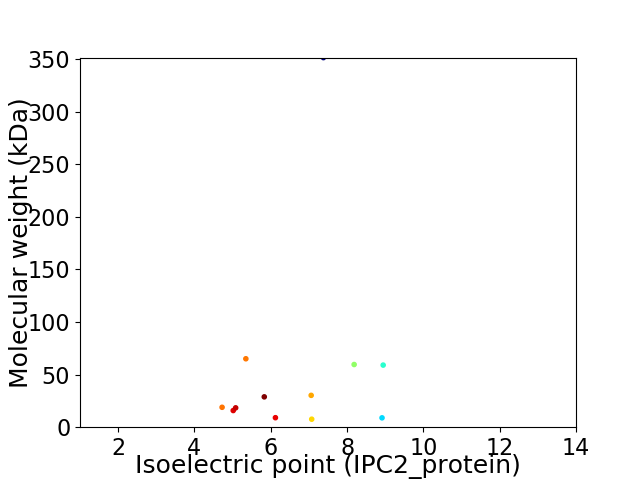
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A8JEL2|A0A0A8JEL2_9CLOS RNA-dependent RNA polymerase (Fragment) OS=Persimmon virus B OX=1493829 GN=RdRp PE=4 SV=1
MM1 pKa = 7.86DD2 pKa = 4.4SKK4 pKa = 11.64VIVTLNLNGSLSAIHH19 pKa = 6.74NSDD22 pKa = 3.48TNVNEE27 pKa = 3.7QSIIEE32 pKa = 4.19SLFIKK37 pKa = 9.99NWFRR41 pKa = 11.84ITDD44 pKa = 3.72DD45 pKa = 3.9LAVEE49 pKa = 4.18EE50 pKa = 4.47TVRR53 pKa = 11.84LLIEE57 pKa = 4.39SEE59 pKa = 4.08SGKK62 pKa = 10.68YY63 pKa = 9.73KK64 pKa = 10.27YY65 pKa = 10.38LSNKK69 pKa = 9.29GCVSEE74 pKa = 5.31LALDD78 pKa = 4.52LNVLLNDD85 pKa = 4.33FGISTGQKK93 pKa = 6.64TVRR96 pKa = 11.84TLVFGSDD103 pKa = 3.19QSNLLKK109 pKa = 11.07SMLISSHH116 pKa = 6.5YY117 pKa = 8.44YY118 pKa = 9.43LSVYY122 pKa = 10.22FLDD125 pKa = 3.74YY126 pKa = 11.29AVFSVVKK133 pKa = 9.76PWEE136 pKa = 4.3CVTNDD141 pKa = 3.06TYY143 pKa = 11.66HH144 pKa = 6.29FVVNNKK150 pKa = 9.1IDD152 pKa = 4.6LEE154 pKa = 4.49DD155 pKa = 4.12DD156 pKa = 3.64CAIIPGMRR164 pKa = 11.84WMCC167 pKa = 3.93
MM1 pKa = 7.86DD2 pKa = 4.4SKK4 pKa = 11.64VIVTLNLNGSLSAIHH19 pKa = 6.74NSDD22 pKa = 3.48TNVNEE27 pKa = 3.7QSIIEE32 pKa = 4.19SLFIKK37 pKa = 9.99NWFRR41 pKa = 11.84ITDD44 pKa = 3.72DD45 pKa = 3.9LAVEE49 pKa = 4.18EE50 pKa = 4.47TVRR53 pKa = 11.84LLIEE57 pKa = 4.39SEE59 pKa = 4.08SGKK62 pKa = 10.68YY63 pKa = 9.73KK64 pKa = 10.27YY65 pKa = 10.38LSNKK69 pKa = 9.29GCVSEE74 pKa = 5.31LALDD78 pKa = 4.52LNVLLNDD85 pKa = 4.33FGISTGQKK93 pKa = 6.64TVRR96 pKa = 11.84TLVFGSDD103 pKa = 3.19QSNLLKK109 pKa = 11.07SMLISSHH116 pKa = 6.5YY117 pKa = 8.44YY118 pKa = 9.43LSVYY122 pKa = 10.22FLDD125 pKa = 3.74YY126 pKa = 11.29AVFSVVKK133 pKa = 9.76PWEE136 pKa = 4.3CVTNDD141 pKa = 3.06TYY143 pKa = 11.66HH144 pKa = 6.29FVVNNKK150 pKa = 9.1IDD152 pKa = 4.6LEE154 pKa = 4.49DD155 pKa = 4.12DD156 pKa = 3.64CAIIPGMRR164 pKa = 11.84WMCC167 pKa = 3.93
Molecular weight: 18.92 kDa
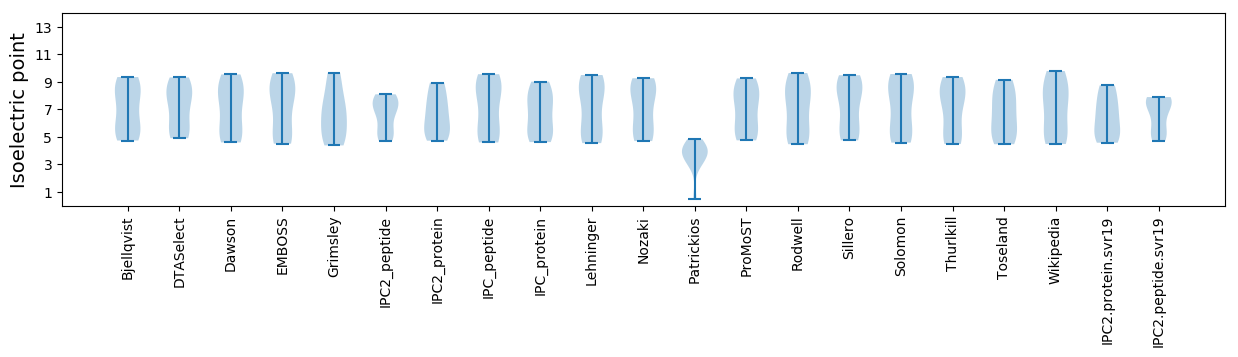
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A8JCX5|A0A0A8JCX5_9CLOS 19 kDa protein OS=Persimmon virus B OX=1493829 GN=p19 PE=4 SV=1
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84AGSMTMPVQTRR14 pKa = 11.84YY15 pKa = 7.48WQPGHH20 pKa = 7.18LFRR23 pKa = 11.84IFFGRR28 pKa = 11.84QDD30 pKa = 2.94IEE32 pKa = 4.81KK33 pKa = 9.83YY34 pKa = 9.78VGEE37 pKa = 5.03AISTYY42 pKa = 8.6YY43 pKa = 10.15NRR45 pKa = 11.84DD46 pKa = 2.99YY47 pKa = 8.32TTQVQYY53 pKa = 11.68VFGDD57 pKa = 3.47SFRR60 pKa = 11.84IITLSPYY67 pKa = 8.54VTSTNSISTAYY78 pKa = 10.54NEE80 pKa = 3.88YY81 pKa = 10.46NLLYY85 pKa = 10.16FYY87 pKa = 10.18VIRR90 pKa = 11.84GGYY93 pKa = 7.28SSKK96 pKa = 10.64VVGYY100 pKa = 9.47QPYY103 pKa = 9.87ILLRR107 pKa = 11.84GILEE111 pKa = 4.1RR112 pKa = 11.84DD113 pKa = 2.92ISTVIMTPLINLSKK127 pKa = 10.94DD128 pKa = 3.64RR129 pKa = 11.84LWANMKK135 pKa = 10.09FKK137 pKa = 11.02SSDD140 pKa = 3.25PRR142 pKa = 11.84FLTLKK147 pKa = 10.85AEE149 pKa = 4.21DD150 pKa = 3.39RR151 pKa = 11.84QFYY154 pKa = 11.04VNLSLVTGEE163 pKa = 4.29FTTSVAKK170 pKa = 9.78TRR172 pKa = 11.84EE173 pKa = 4.27AKK175 pKa = 10.42DD176 pKa = 3.62VFPDD180 pKa = 3.44IPVSEE185 pKa = 4.61SVSSGLPLKK194 pKa = 10.84YY195 pKa = 10.01ILPKK199 pKa = 10.75LFFHH203 pKa = 7.79GIRR206 pKa = 11.84VLNGNVATKK215 pKa = 10.49LYY217 pKa = 9.59VQYY220 pKa = 11.27NHH222 pKa = 5.98VLAVLSNILSSYY234 pKa = 10.15NHH236 pKa = 6.73LFMLNSSFTHH246 pKa = 5.07YY247 pKa = 10.9AITFFGWMAMFFDD260 pKa = 5.49NYY262 pKa = 10.93SQLNYY267 pKa = 10.51EE268 pKa = 4.55SGEE271 pKa = 3.82KK272 pKa = 9.25WEE274 pKa = 4.13EE275 pKa = 3.67VMRR278 pKa = 11.84FCTHH282 pKa = 7.38PLTSFFIKK290 pKa = 10.49FDD292 pKa = 4.17DD293 pKa = 4.52SDD295 pKa = 3.7LQRR298 pKa = 11.84IRR300 pKa = 11.84WITPDD305 pKa = 3.13RR306 pKa = 11.84FEE308 pKa = 5.94KK309 pKa = 10.13VLQYY313 pKa = 11.4VNYY316 pKa = 9.93VSYY319 pKa = 11.23GGLALSDD326 pKa = 3.91YY327 pKa = 11.12QEE329 pKa = 5.51LIWTPKK335 pKa = 10.46VYY337 pKa = 10.5DD338 pKa = 4.16KK339 pKa = 11.57EE340 pKa = 4.7SFCQLSAIFVSLVSTNKK357 pKa = 9.63RR358 pKa = 11.84VEE360 pKa = 4.53PILILGAIIAYY371 pKa = 9.16YY372 pKa = 9.82SIHH375 pKa = 5.09GTCRR379 pKa = 11.84YY380 pKa = 9.7RR381 pKa = 11.84SEE383 pKa = 3.58KK384 pKa = 9.59RR385 pKa = 11.84PRR387 pKa = 11.84YY388 pKa = 9.42FKK390 pKa = 10.43FYY392 pKa = 10.52SHH394 pKa = 6.88GEE396 pKa = 4.02EE397 pKa = 4.01IVCDD401 pKa = 4.88FKK403 pKa = 11.38FLEE406 pKa = 4.4IKK408 pKa = 10.32FDD410 pKa = 3.43MMQNFVKK417 pKa = 10.12AYY419 pKa = 7.9SVRR422 pKa = 11.84RR423 pKa = 11.84VYY425 pKa = 10.72LGSAGDD431 pKa = 3.66LCVDD435 pKa = 4.27FLRR438 pKa = 11.84SIGLEE443 pKa = 3.76LPLRR447 pKa = 11.84WKK449 pKa = 10.21RR450 pKa = 11.84VNWLPYY456 pKa = 9.67KK457 pKa = 10.09DD458 pKa = 3.72VRR460 pKa = 11.84YY461 pKa = 10.2VDD463 pKa = 3.55YY464 pKa = 10.75HH465 pKa = 6.67KK466 pKa = 10.47HH467 pKa = 3.83VTNKK471 pKa = 9.98NMLKK475 pKa = 10.24KK476 pKa = 10.54GIRR479 pKa = 11.84ASIGAYY485 pKa = 9.99GNVVSTNAFEE495 pKa = 4.0ARR497 pKa = 11.84KK498 pKa = 8.88WRR500 pKa = 11.84DD501 pKa = 2.7LWW503 pKa = 5.05
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84AGSMTMPVQTRR14 pKa = 11.84YY15 pKa = 7.48WQPGHH20 pKa = 7.18LFRR23 pKa = 11.84IFFGRR28 pKa = 11.84QDD30 pKa = 2.94IEE32 pKa = 4.81KK33 pKa = 9.83YY34 pKa = 9.78VGEE37 pKa = 5.03AISTYY42 pKa = 8.6YY43 pKa = 10.15NRR45 pKa = 11.84DD46 pKa = 2.99YY47 pKa = 8.32TTQVQYY53 pKa = 11.68VFGDD57 pKa = 3.47SFRR60 pKa = 11.84IITLSPYY67 pKa = 8.54VTSTNSISTAYY78 pKa = 10.54NEE80 pKa = 3.88YY81 pKa = 10.46NLLYY85 pKa = 10.16FYY87 pKa = 10.18VIRR90 pKa = 11.84GGYY93 pKa = 7.28SSKK96 pKa = 10.64VVGYY100 pKa = 9.47QPYY103 pKa = 9.87ILLRR107 pKa = 11.84GILEE111 pKa = 4.1RR112 pKa = 11.84DD113 pKa = 2.92ISTVIMTPLINLSKK127 pKa = 10.94DD128 pKa = 3.64RR129 pKa = 11.84LWANMKK135 pKa = 10.09FKK137 pKa = 11.02SSDD140 pKa = 3.25PRR142 pKa = 11.84FLTLKK147 pKa = 10.85AEE149 pKa = 4.21DD150 pKa = 3.39RR151 pKa = 11.84QFYY154 pKa = 11.04VNLSLVTGEE163 pKa = 4.29FTTSVAKK170 pKa = 9.78TRR172 pKa = 11.84EE173 pKa = 4.27AKK175 pKa = 10.42DD176 pKa = 3.62VFPDD180 pKa = 3.44IPVSEE185 pKa = 4.61SVSSGLPLKK194 pKa = 10.84YY195 pKa = 10.01ILPKK199 pKa = 10.75LFFHH203 pKa = 7.79GIRR206 pKa = 11.84VLNGNVATKK215 pKa = 10.49LYY217 pKa = 9.59VQYY220 pKa = 11.27NHH222 pKa = 5.98VLAVLSNILSSYY234 pKa = 10.15NHH236 pKa = 6.73LFMLNSSFTHH246 pKa = 5.07YY247 pKa = 10.9AITFFGWMAMFFDD260 pKa = 5.49NYY262 pKa = 10.93SQLNYY267 pKa = 10.51EE268 pKa = 4.55SGEE271 pKa = 3.82KK272 pKa = 9.25WEE274 pKa = 4.13EE275 pKa = 3.67VMRR278 pKa = 11.84FCTHH282 pKa = 7.38PLTSFFIKK290 pKa = 10.49FDD292 pKa = 4.17DD293 pKa = 4.52SDD295 pKa = 3.7LQRR298 pKa = 11.84IRR300 pKa = 11.84WITPDD305 pKa = 3.13RR306 pKa = 11.84FEE308 pKa = 5.94KK309 pKa = 10.13VLQYY313 pKa = 11.4VNYY316 pKa = 9.93VSYY319 pKa = 11.23GGLALSDD326 pKa = 3.91YY327 pKa = 11.12QEE329 pKa = 5.51LIWTPKK335 pKa = 10.46VYY337 pKa = 10.5DD338 pKa = 4.16KK339 pKa = 11.57EE340 pKa = 4.7SFCQLSAIFVSLVSTNKK357 pKa = 9.63RR358 pKa = 11.84VEE360 pKa = 4.53PILILGAIIAYY371 pKa = 9.16YY372 pKa = 9.82SIHH375 pKa = 5.09GTCRR379 pKa = 11.84YY380 pKa = 9.7RR381 pKa = 11.84SEE383 pKa = 3.58KK384 pKa = 9.59RR385 pKa = 11.84PRR387 pKa = 11.84YY388 pKa = 9.42FKK390 pKa = 10.43FYY392 pKa = 10.52SHH394 pKa = 6.88GEE396 pKa = 4.02EE397 pKa = 4.01IVCDD401 pKa = 4.88FKK403 pKa = 11.38FLEE406 pKa = 4.4IKK408 pKa = 10.32FDD410 pKa = 3.43MMQNFVKK417 pKa = 10.12AYY419 pKa = 7.9SVRR422 pKa = 11.84RR423 pKa = 11.84VYY425 pKa = 10.72LGSAGDD431 pKa = 3.66LCVDD435 pKa = 4.27FLRR438 pKa = 11.84SIGLEE443 pKa = 3.76LPLRR447 pKa = 11.84WKK449 pKa = 10.21RR450 pKa = 11.84VNWLPYY456 pKa = 9.67KK457 pKa = 10.09DD458 pKa = 3.72VRR460 pKa = 11.84YY461 pKa = 10.2VDD463 pKa = 3.55YY464 pKa = 10.75HH465 pKa = 6.67KK466 pKa = 10.47HH467 pKa = 3.83VTNKK471 pKa = 9.98NMLKK475 pKa = 10.24KK476 pKa = 10.54GIRR479 pKa = 11.84ASIGAYY485 pKa = 9.99GNVVSTNAFEE495 pKa = 4.0ARR497 pKa = 11.84KK498 pKa = 8.88WRR500 pKa = 11.84DD501 pKa = 2.7LWW503 pKa = 5.05
Molecular weight: 59.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5961 |
69 |
3149 |
496.8 |
56.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.207 ± 0.523 | 2.399 ± 0.341 |
5.838 ± 0.343 | 5.955 ± 0.481 |
4.546 ± 0.455 | 5.871 ± 0.498 |
1.627 ± 0.182 | 5.788 ± 0.659 |
6.123 ± 0.243 | 8.757 ± 0.468 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.298 ± 0.216 | 4.462 ± 0.706 |
3.942 ± 0.693 | 2.214 ± 0.136 |
6.123 ± 0.479 | 8.489 ± 0.313 |
4.815 ± 0.431 | 8.824 ± 0.433 |
1.174 ± 0.185 | 4.546 ± 0.698 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
