
Mycoplasma phage phiMFV1
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
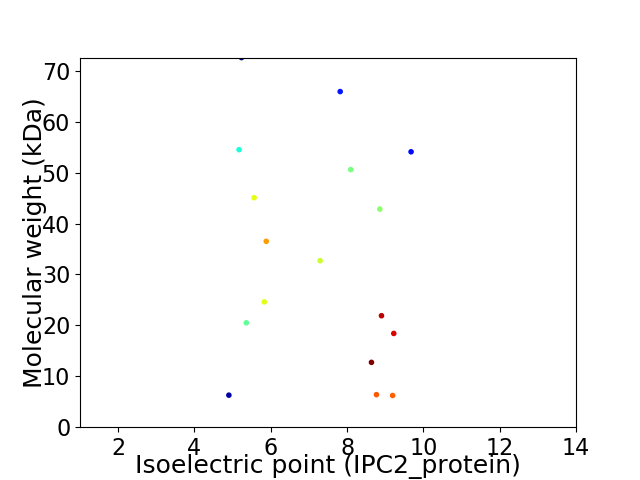
Virtual 2D-PAGE plot for 17 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|Q6GYZ6|Q6GYZ6_9VIRU ABC transmembrane type-1 domain-containing protein (Fragment) OS=Mycoplasma phage phiMFV1 OX=280702 PE=4 SV=1
MM1 pKa = 7.7LNNKK5 pKa = 9.43CDD7 pKa = 3.68FCHH10 pKa = 7.42KK11 pKa = 10.17RR12 pKa = 11.84FQPKK16 pKa = 9.45KK17 pKa = 10.15EE18 pKa = 3.8RR19 pKa = 11.84YY20 pKa = 7.88FWGNLVVCSEE30 pKa = 4.28EE31 pKa = 4.36CLEE34 pKa = 4.41QVSCYY39 pKa = 10.62ASEE42 pKa = 4.19IDD44 pKa = 4.8LNCEE48 pKa = 4.0GEE50 pKa = 4.19NNEE53 pKa = 4.24
MM1 pKa = 7.7LNNKK5 pKa = 9.43CDD7 pKa = 3.68FCHH10 pKa = 7.42KK11 pKa = 10.17RR12 pKa = 11.84FQPKK16 pKa = 9.45KK17 pKa = 10.15EE18 pKa = 3.8RR19 pKa = 11.84YY20 pKa = 7.88FWGNLVVCSEE30 pKa = 4.28EE31 pKa = 4.36CLEE34 pKa = 4.41QVSCYY39 pKa = 10.62ASEE42 pKa = 4.19IDD44 pKa = 4.8LNCEE48 pKa = 4.0GEE50 pKa = 4.19NNEE53 pKa = 4.24
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5QGJ7|Q5QGJ7_9VIRU HtpE OS=Mycoplasma phage phiMFV1 OX=280702 GN=htpE PE=4 SV=1
MM1 pKa = 7.75LIYY4 pKa = 10.73SNPKK8 pKa = 9.77PKK10 pKa = 8.35TTLQLSPKK18 pKa = 9.94SFVINDD24 pKa = 3.72SKK26 pKa = 11.57KK27 pKa = 10.37QMNPFLRR34 pKa = 11.84GFLSFLHH41 pKa = 5.85VALDD45 pKa = 3.58IGASFLGSFVNTFAPGLGLVIEE67 pKa = 4.84YY68 pKa = 10.61GLSTALDD75 pKa = 3.93LGFNAAINYY84 pKa = 9.23GNINWQDD91 pKa = 3.48FGISAGINAIPFVTRR106 pKa = 11.84GIRR109 pKa = 11.84YY110 pKa = 8.16AANTHH115 pKa = 5.19KK116 pKa = 9.74TNKK119 pKa = 9.34FFQLAKK125 pKa = 9.87NLKK128 pKa = 10.14KK129 pKa = 10.57EE130 pKa = 4.16GQEE133 pKa = 3.9EE134 pKa = 4.24LSEE137 pKa = 4.77KK138 pKa = 8.1ITKK141 pKa = 9.63IAANYY146 pKa = 10.12SEE148 pKa = 5.48LGVDD152 pKa = 3.03KK153 pKa = 11.23AKK155 pKa = 10.97FIDD158 pKa = 3.84QKK160 pKa = 11.72SLLKK164 pKa = 10.66KK165 pKa = 9.73EE166 pKa = 4.58LKK168 pKa = 10.59LFNNDD173 pKa = 2.37SFLRR177 pKa = 11.84TLSDD181 pKa = 3.27NKK183 pKa = 10.54NIYY186 pKa = 10.25YY187 pKa = 10.28FLNKK191 pKa = 9.47KK192 pKa = 7.82VQNIQKK198 pKa = 10.15AFTHH202 pKa = 5.61LRR204 pKa = 11.84SMVSLLTSPSYY215 pKa = 10.23LAKK218 pKa = 10.51KK219 pKa = 10.44AVDD222 pKa = 3.59TVFKK226 pKa = 10.77HH227 pKa = 5.72PKK229 pKa = 10.11AYY231 pKa = 10.6LNKK234 pKa = 9.92QLGKK238 pKa = 8.76FTKK241 pKa = 10.25LLKK244 pKa = 9.61NTFKK248 pKa = 10.91SKK250 pKa = 10.24PIKK253 pKa = 10.37KK254 pKa = 9.27VVKK257 pKa = 10.05KK258 pKa = 10.0ASNYY262 pKa = 8.55IWIDD266 pKa = 3.06HH267 pKa = 5.81SSWLEE272 pKa = 3.48RR273 pKa = 11.84MSINKK278 pKa = 8.7VNNPWDD284 pKa = 3.31IKK286 pKa = 10.93AINAVLYY293 pKa = 8.99FKK295 pKa = 11.16KK296 pKa = 10.48NATKK300 pKa = 10.49SKK302 pKa = 10.22KK303 pKa = 9.23NPKK306 pKa = 9.45GKK308 pKa = 10.41EE309 pKa = 3.76PVRR312 pKa = 11.84LWNKK316 pKa = 10.63SIDD319 pKa = 3.81EE320 pKa = 4.7LIKK323 pKa = 10.48LVCAKK328 pKa = 10.74SPGRR332 pKa = 11.84YY333 pKa = 8.67YY334 pKa = 11.05LNNIAYY340 pKa = 8.23GHH342 pKa = 5.62IVGRR346 pKa = 11.84FIRR349 pKa = 11.84KK350 pKa = 9.02FEE352 pKa = 3.97MFQAVPIHH360 pKa = 6.62PIFSNLIGTSLFTFRR375 pKa = 11.84TINSLIMNFKK385 pKa = 7.92RR386 pKa = 11.84QKK388 pKa = 9.49WVWLEE393 pKa = 3.95DD394 pKa = 3.29KK395 pKa = 11.04NIILNEE401 pKa = 3.68IVKK404 pKa = 10.0GVKK407 pKa = 10.1EE408 pKa = 4.02NVFKK412 pKa = 10.75GWRR415 pKa = 11.84APGFQFISSMARR427 pKa = 11.84SWATGNVGYY436 pKa = 7.45QTKK439 pKa = 10.43AALRR443 pKa = 11.84WANKK447 pKa = 9.58KK448 pKa = 10.56AFTSKK453 pKa = 7.84TQKK456 pKa = 8.36TNYY459 pKa = 8.17WVNHH463 pKa = 4.26WVKK466 pKa = 10.94NKK468 pKa = 10.01YY469 pKa = 9.78KK470 pKa = 10.57KK471 pKa = 10.53
MM1 pKa = 7.75LIYY4 pKa = 10.73SNPKK8 pKa = 9.77PKK10 pKa = 8.35TTLQLSPKK18 pKa = 9.94SFVINDD24 pKa = 3.72SKK26 pKa = 11.57KK27 pKa = 10.37QMNPFLRR34 pKa = 11.84GFLSFLHH41 pKa = 5.85VALDD45 pKa = 3.58IGASFLGSFVNTFAPGLGLVIEE67 pKa = 4.84YY68 pKa = 10.61GLSTALDD75 pKa = 3.93LGFNAAINYY84 pKa = 9.23GNINWQDD91 pKa = 3.48FGISAGINAIPFVTRR106 pKa = 11.84GIRR109 pKa = 11.84YY110 pKa = 8.16AANTHH115 pKa = 5.19KK116 pKa = 9.74TNKK119 pKa = 9.34FFQLAKK125 pKa = 9.87NLKK128 pKa = 10.14KK129 pKa = 10.57EE130 pKa = 4.16GQEE133 pKa = 3.9EE134 pKa = 4.24LSEE137 pKa = 4.77KK138 pKa = 8.1ITKK141 pKa = 9.63IAANYY146 pKa = 10.12SEE148 pKa = 5.48LGVDD152 pKa = 3.03KK153 pKa = 11.23AKK155 pKa = 10.97FIDD158 pKa = 3.84QKK160 pKa = 11.72SLLKK164 pKa = 10.66KK165 pKa = 9.73EE166 pKa = 4.58LKK168 pKa = 10.59LFNNDD173 pKa = 2.37SFLRR177 pKa = 11.84TLSDD181 pKa = 3.27NKK183 pKa = 10.54NIYY186 pKa = 10.25YY187 pKa = 10.28FLNKK191 pKa = 9.47KK192 pKa = 7.82VQNIQKK198 pKa = 10.15AFTHH202 pKa = 5.61LRR204 pKa = 11.84SMVSLLTSPSYY215 pKa = 10.23LAKK218 pKa = 10.51KK219 pKa = 10.44AVDD222 pKa = 3.59TVFKK226 pKa = 10.77HH227 pKa = 5.72PKK229 pKa = 10.11AYY231 pKa = 10.6LNKK234 pKa = 9.92QLGKK238 pKa = 8.76FTKK241 pKa = 10.25LLKK244 pKa = 9.61NTFKK248 pKa = 10.91SKK250 pKa = 10.24PIKK253 pKa = 10.37KK254 pKa = 9.27VVKK257 pKa = 10.05KK258 pKa = 10.0ASNYY262 pKa = 8.55IWIDD266 pKa = 3.06HH267 pKa = 5.81SSWLEE272 pKa = 3.48RR273 pKa = 11.84MSINKK278 pKa = 8.7VNNPWDD284 pKa = 3.31IKK286 pKa = 10.93AINAVLYY293 pKa = 8.99FKK295 pKa = 11.16KK296 pKa = 10.48NATKK300 pKa = 10.49SKK302 pKa = 10.22KK303 pKa = 9.23NPKK306 pKa = 9.45GKK308 pKa = 10.41EE309 pKa = 3.76PVRR312 pKa = 11.84LWNKK316 pKa = 10.63SIDD319 pKa = 3.81EE320 pKa = 4.7LIKK323 pKa = 10.48LVCAKK328 pKa = 10.74SPGRR332 pKa = 11.84YY333 pKa = 8.67YY334 pKa = 11.05LNNIAYY340 pKa = 8.23GHH342 pKa = 5.62IVGRR346 pKa = 11.84FIRR349 pKa = 11.84KK350 pKa = 9.02FEE352 pKa = 3.97MFQAVPIHH360 pKa = 6.62PIFSNLIGTSLFTFRR375 pKa = 11.84TINSLIMNFKK385 pKa = 7.92RR386 pKa = 11.84QKK388 pKa = 9.49WVWLEE393 pKa = 3.95DD394 pKa = 3.29KK395 pKa = 11.04NIILNEE401 pKa = 3.68IVKK404 pKa = 10.0GVKK407 pKa = 10.1EE408 pKa = 4.02NVFKK412 pKa = 10.75GWRR415 pKa = 11.84APGFQFISSMARR427 pKa = 11.84SWATGNVGYY436 pKa = 7.45QTKK439 pKa = 10.43AALRR443 pKa = 11.84WANKK447 pKa = 9.58KK448 pKa = 10.56AFTSKK453 pKa = 7.84TQKK456 pKa = 8.36TNYY459 pKa = 8.17WVNHH463 pKa = 4.26WVKK466 pKa = 10.94NKK468 pKa = 10.01YY469 pKa = 9.78KK470 pKa = 10.57KK471 pKa = 10.53
Molecular weight: 54.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4901 |
53 |
615 |
288.3 |
33.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.264 ± 0.344 | 0.673 ± 0.174 |
4.713 ± 0.351 | 7.631 ± 0.558 |
6.06 ± 0.37 | 3.632 ± 0.243 |
1.041 ± 0.178 | 8.835 ± 0.443 |
11.589 ± 0.598 | 10.161 ± 0.496 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.673 ± 0.108 | 9.529 ± 0.537 |
2.571 ± 0.27 | 3.836 ± 0.236 |
2.346 ± 0.238 | 6.305 ± 0.476 |
4.652 ± 0.28 | 4.55 ± 0.312 |
1.51 ± 0.187 | 4.428 ± 0.288 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
