
Trinickia dinghuensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Trinickia
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
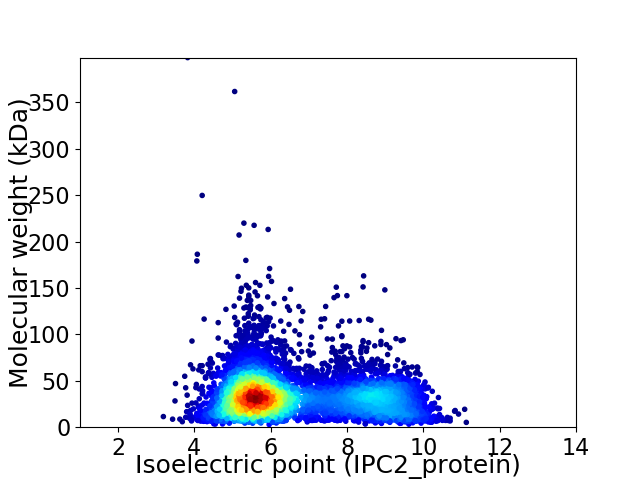
Virtual 2D-PAGE plot for 6498 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
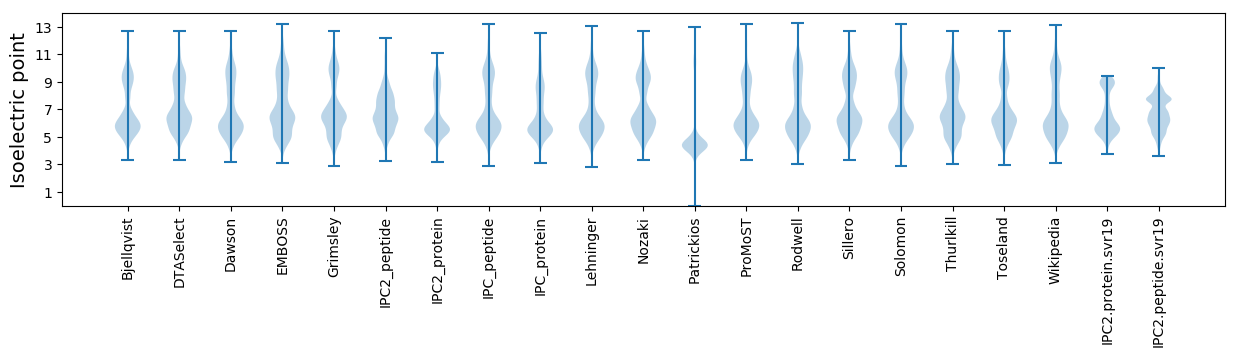
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3D8JQC6|A0A3D8JQC6_9BURK Protein translocase subunit SecD OS=Trinickia dinghuensis OX=2291023 GN=secD PE=3 SV=1
MM1 pKa = 7.5KK2 pKa = 10.23LAASLSKK9 pKa = 10.8LFAAMRR15 pKa = 11.84ASEE18 pKa = 4.31TSGRR22 pKa = 11.84TQYY25 pKa = 11.54AAVPAPLIVALEE37 pKa = 3.82PRR39 pKa = 11.84IVYY42 pKa = 9.28DD43 pKa = 4.48ASAASIGMAAAVAAQHH59 pKa = 5.67HH60 pKa = 6.32HH61 pKa = 6.65ASAEE65 pKa = 4.03AHH67 pKa = 6.68DD68 pKa = 4.79AATPAAVSNSAASAGRR84 pKa = 11.84SAAFSVSSSDD94 pKa = 3.16AAASQRR100 pKa = 11.84AVHH103 pKa = 6.91GYY105 pKa = 7.26MTAQSSTGIGAGDD118 pKa = 3.99TSTMRR123 pKa = 11.84AVHH126 pKa = 7.03GYY128 pKa = 8.23TDD130 pKa = 4.25LAVTSADD137 pKa = 3.46KK138 pKa = 10.97QVVFVNSNVTDD149 pKa = 3.85YY150 pKa = 11.02QALIAGVPQGTQVVILDD167 pKa = 3.74STKK170 pKa = 11.11DD171 pKa = 3.47GLSQMEE177 pKa = 4.68QYY179 pKa = 10.23LQQHH183 pKa = 6.71PGVSAIHH190 pKa = 5.92LVSHH194 pKa = 6.63GADD197 pKa = 3.1GDD199 pKa = 4.1FEE201 pKa = 5.64IGSTWINEE209 pKa = 4.2ADD211 pKa = 3.64LSTYY215 pKa = 10.18SAQLAQIGAAMKK227 pKa = 10.27PGGDD231 pKa = 3.67FLIYY235 pKa = 10.61GCDD238 pKa = 3.26VAEE241 pKa = 4.39NADD244 pKa = 3.55GKK246 pKa = 11.22ALVQQIASITGLNVAASTDD265 pKa = 3.21ITGAASLGGDD275 pKa = 3.06WTLEE279 pKa = 3.76YY280 pKa = 10.96DD281 pKa = 3.84VGNVHH286 pKa = 6.46TNVIFSAAAEE296 pKa = 4.0KK297 pKa = 10.81SYY299 pKa = 11.07DD300 pKa = 3.59YY301 pKa = 10.92TLALIDD307 pKa = 4.22EE308 pKa = 5.24NYY310 pKa = 10.17DD311 pKa = 3.32SHH313 pKa = 9.13VGFDD317 pKa = 3.36TGGTGVSSTTLDD329 pKa = 3.13GLTYY333 pKa = 10.56TGDD336 pKa = 3.44QPVEE340 pKa = 4.01YY341 pKa = 9.74QVVSGSLPNSATGGEE356 pKa = 4.32LVVNTQGTAMSTMTVSRR373 pKa = 11.84ADD375 pKa = 3.49GSLMAVQSFDD385 pKa = 3.38IDD387 pKa = 3.19IFLSNDD393 pKa = 3.01VTVQAIGSSGQVLGSIDD410 pKa = 5.01LNLNDD415 pKa = 4.16GASTNTYY422 pKa = 10.56DD423 pKa = 4.17AGHH426 pKa = 6.45SSGTSLFHH434 pKa = 7.62VNLTSLAAFSQVKK447 pKa = 9.87SIKK450 pKa = 10.42FIEE453 pKa = 4.16NSGNGTYY460 pKa = 10.89LSPTLDD466 pKa = 3.1NFTYY470 pKa = 10.83LDD472 pKa = 3.77PTSPPTLTASGGTASFTAADD492 pKa = 4.0NAASTPVVVDD502 pKa = 3.65PGITLTDD509 pKa = 3.91GNSATATSATVTITGNFQAGEE530 pKa = 4.02DD531 pKa = 3.44QLAFINNNSTTYY543 pKa = 11.26GNIANGSYY551 pKa = 10.34NPSSGVLTLNGTASITQWQAALSAVTYY578 pKa = 9.57TDD580 pKa = 3.19TAVTPNTAMRR590 pKa = 11.84TVSFEE595 pKa = 3.98LTSDD599 pKa = 3.51GFTPSNTVTRR609 pKa = 11.84TVTVADD615 pKa = 3.82TDD617 pKa = 3.55QTPIVHH623 pKa = 5.5TTGGTTSYY631 pKa = 11.76ADD633 pKa = 3.29GTSAVTIDD641 pKa = 3.55SGVTVTDD648 pKa = 4.79LDD650 pKa = 3.75NTTQASGTVSISAGFQSGDD669 pKa = 3.5TLSFTANSSTMGNIAIQSYY688 pKa = 10.58NSATGVLSLTSASALATKK706 pKa = 9.74AQWAAALSAVKK717 pKa = 10.47FSSTSTTYY725 pKa = 11.2GNRR728 pKa = 11.84TISFTTNDD736 pKa = 3.44GTQTSAAATDD746 pKa = 3.82TVDD749 pKa = 4.15VVNPLQLTTDD759 pKa = 3.34SGSAAFVAGDD769 pKa = 3.84NVTSTPVAVDD779 pKa = 3.17SGLTLTDD786 pKa = 3.42AATGTLSSVTVAITGNFDD804 pKa = 3.34SSHH807 pKa = 6.98DD808 pKa = 3.58QLQFTNTGSITGSYY822 pKa = 10.6SSATGVLTLTGASATLTQWQSALQSVTFEE851 pKa = 4.21NTAVTPSNVPRR862 pKa = 11.84TVTFTAVDD870 pKa = 3.67SLTNTSTATRR880 pKa = 11.84TVTVTDD886 pKa = 3.55TDD888 pKa = 3.38QTPIVTTTGGTTNYY902 pKa = 10.22VGGTSGVTIDD912 pKa = 4.26SGVGG916 pKa = 3.08
MM1 pKa = 7.5KK2 pKa = 10.23LAASLSKK9 pKa = 10.8LFAAMRR15 pKa = 11.84ASEE18 pKa = 4.31TSGRR22 pKa = 11.84TQYY25 pKa = 11.54AAVPAPLIVALEE37 pKa = 3.82PRR39 pKa = 11.84IVYY42 pKa = 9.28DD43 pKa = 4.48ASAASIGMAAAVAAQHH59 pKa = 5.67HH60 pKa = 6.32HH61 pKa = 6.65ASAEE65 pKa = 4.03AHH67 pKa = 6.68DD68 pKa = 4.79AATPAAVSNSAASAGRR84 pKa = 11.84SAAFSVSSSDD94 pKa = 3.16AAASQRR100 pKa = 11.84AVHH103 pKa = 6.91GYY105 pKa = 7.26MTAQSSTGIGAGDD118 pKa = 3.99TSTMRR123 pKa = 11.84AVHH126 pKa = 7.03GYY128 pKa = 8.23TDD130 pKa = 4.25LAVTSADD137 pKa = 3.46KK138 pKa = 10.97QVVFVNSNVTDD149 pKa = 3.85YY150 pKa = 11.02QALIAGVPQGTQVVILDD167 pKa = 3.74STKK170 pKa = 11.11DD171 pKa = 3.47GLSQMEE177 pKa = 4.68QYY179 pKa = 10.23LQQHH183 pKa = 6.71PGVSAIHH190 pKa = 5.92LVSHH194 pKa = 6.63GADD197 pKa = 3.1GDD199 pKa = 4.1FEE201 pKa = 5.64IGSTWINEE209 pKa = 4.2ADD211 pKa = 3.64LSTYY215 pKa = 10.18SAQLAQIGAAMKK227 pKa = 10.27PGGDD231 pKa = 3.67FLIYY235 pKa = 10.61GCDD238 pKa = 3.26VAEE241 pKa = 4.39NADD244 pKa = 3.55GKK246 pKa = 11.22ALVQQIASITGLNVAASTDD265 pKa = 3.21ITGAASLGGDD275 pKa = 3.06WTLEE279 pKa = 3.76YY280 pKa = 10.96DD281 pKa = 3.84VGNVHH286 pKa = 6.46TNVIFSAAAEE296 pKa = 4.0KK297 pKa = 10.81SYY299 pKa = 11.07DD300 pKa = 3.59YY301 pKa = 10.92TLALIDD307 pKa = 4.22EE308 pKa = 5.24NYY310 pKa = 10.17DD311 pKa = 3.32SHH313 pKa = 9.13VGFDD317 pKa = 3.36TGGTGVSSTTLDD329 pKa = 3.13GLTYY333 pKa = 10.56TGDD336 pKa = 3.44QPVEE340 pKa = 4.01YY341 pKa = 9.74QVVSGSLPNSATGGEE356 pKa = 4.32LVVNTQGTAMSTMTVSRR373 pKa = 11.84ADD375 pKa = 3.49GSLMAVQSFDD385 pKa = 3.38IDD387 pKa = 3.19IFLSNDD393 pKa = 3.01VTVQAIGSSGQVLGSIDD410 pKa = 5.01LNLNDD415 pKa = 4.16GASTNTYY422 pKa = 10.56DD423 pKa = 4.17AGHH426 pKa = 6.45SSGTSLFHH434 pKa = 7.62VNLTSLAAFSQVKK447 pKa = 9.87SIKK450 pKa = 10.42FIEE453 pKa = 4.16NSGNGTYY460 pKa = 10.89LSPTLDD466 pKa = 3.1NFTYY470 pKa = 10.83LDD472 pKa = 3.77PTSPPTLTASGGTASFTAADD492 pKa = 4.0NAASTPVVVDD502 pKa = 3.65PGITLTDD509 pKa = 3.91GNSATATSATVTITGNFQAGEE530 pKa = 4.02DD531 pKa = 3.44QLAFINNNSTTYY543 pKa = 11.26GNIANGSYY551 pKa = 10.34NPSSGVLTLNGTASITQWQAALSAVTYY578 pKa = 9.57TDD580 pKa = 3.19TAVTPNTAMRR590 pKa = 11.84TVSFEE595 pKa = 3.98LTSDD599 pKa = 3.51GFTPSNTVTRR609 pKa = 11.84TVTVADD615 pKa = 3.82TDD617 pKa = 3.55QTPIVHH623 pKa = 5.5TTGGTTSYY631 pKa = 11.76ADD633 pKa = 3.29GTSAVTIDD641 pKa = 3.55SGVTVTDD648 pKa = 4.79LDD650 pKa = 3.75NTTQASGTVSISAGFQSGDD669 pKa = 3.5TLSFTANSSTMGNIAIQSYY688 pKa = 10.58NSATGVLSLTSASALATKK706 pKa = 9.74AQWAAALSAVKK717 pKa = 10.47FSSTSTTYY725 pKa = 11.2GNRR728 pKa = 11.84TISFTTNDD736 pKa = 3.44GTQTSAAATDD746 pKa = 3.82TVDD749 pKa = 4.15VVNPLQLTTDD759 pKa = 3.34SGSAAFVAGDD769 pKa = 3.84NVTSTPVAVDD779 pKa = 3.17SGLTLTDD786 pKa = 3.42AATGTLSSVTVAITGNFDD804 pKa = 3.34SSHH807 pKa = 6.98DD808 pKa = 3.58QLQFTNTGSITGSYY822 pKa = 10.6SSATGVLTLTGASATLTQWQSALQSVTFEE851 pKa = 4.21NTAVTPSNVPRR862 pKa = 11.84TVTFTAVDD870 pKa = 3.67SLTNTSTATRR880 pKa = 11.84TVTVTDD886 pKa = 3.55TDD888 pKa = 3.38QTPIVTTTGGTTNYY902 pKa = 10.22VGGTSGVTIDD912 pKa = 4.26SGVGG916 pKa = 3.08
Molecular weight: 92.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3D8K1U8|A0A3D8K1U8_9BURK Acetolactate synthase large subunit OS=Trinickia dinghuensis OX=2291023 GN=DWV00_11390 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.06QPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LAII44 pKa = 4.0
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.06QPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LAII44 pKa = 4.0
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2122929 |
24 |
3980 |
326.7 |
35.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.839 ± 0.042 | 0.959 ± 0.011 |
5.435 ± 0.022 | 5.274 ± 0.034 |
3.674 ± 0.019 | 8.213 ± 0.034 |
2.33 ± 0.015 | 4.688 ± 0.022 |
3.027 ± 0.024 | 10.075 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.346 ± 0.015 | 2.736 ± 0.019 |
5.064 ± 0.024 | 3.48 ± 0.022 |
7.039 ± 0.038 | 5.9 ± 0.033 |
5.447 ± 0.032 | 7.691 ± 0.024 |
1.341 ± 0.013 | 2.443 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
