
Cellulomonas carbonis T26
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas; Cellulomonas carbonis
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
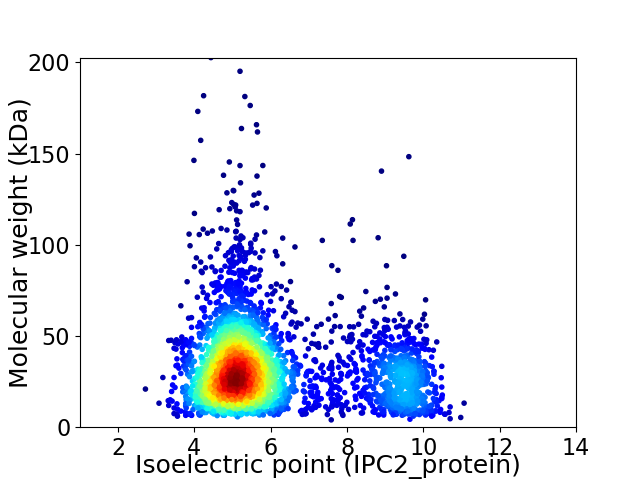
Virtual 2D-PAGE plot for 3407 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
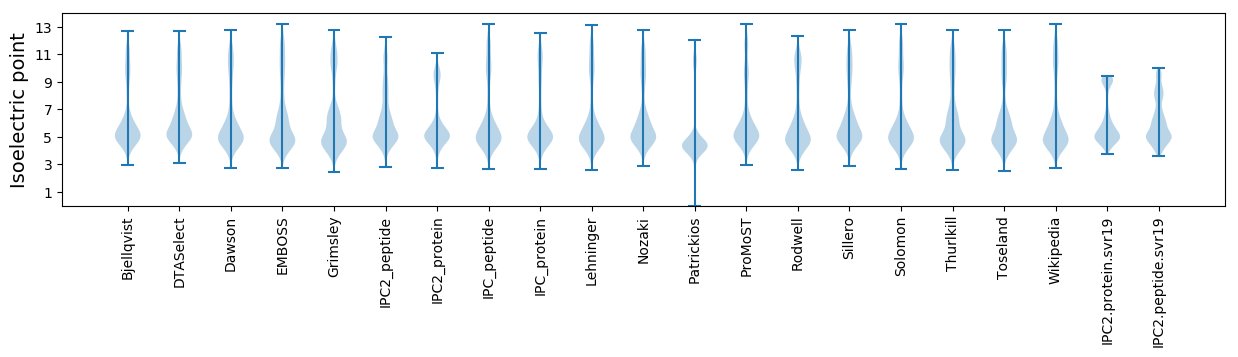
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0BXQ5|A0A0A0BXQ5_9CELL Chemotaxis protein CheY OS=Cellulomonas carbonis T26 OX=947969 GN=N868_11905 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.61MRR4 pKa = 11.84RR5 pKa = 11.84IGAAGAIVATGALALSACSSTPSTTTEE32 pKa = 3.61IDD34 pKa = 3.41EE35 pKa = 4.35EE36 pKa = 4.61TSLSIGWNQPFYY48 pKa = 10.99SQNNLTSVGNATTNSNVIYY67 pKa = 9.01LTNAAFNYY75 pKa = 9.87YY76 pKa = 10.68DD77 pKa = 4.31NNLEE81 pKa = 4.09LQQFSDD87 pKa = 3.51FGTYY91 pKa = 7.97EE92 pKa = 4.08VEE94 pKa = 4.61SEE96 pKa = 4.46DD97 pKa = 4.36PLTVTYY103 pKa = 9.05TINEE107 pKa = 4.16GATWSDD113 pKa = 3.52GTPVDD118 pKa = 3.54AVDD121 pKa = 3.75VFLYY125 pKa = 9.92WGAQNDD131 pKa = 4.03SFDD134 pKa = 3.95NVAPEE139 pKa = 3.8YY140 pKa = 10.76DD141 pKa = 3.58EE142 pKa = 5.14EE143 pKa = 5.35GNITNQDD150 pKa = 3.7AIDD153 pKa = 3.75AGVYY157 pKa = 10.11FDD159 pKa = 4.39SSSVAMNLISEE170 pKa = 4.83PPTISDD176 pKa = 4.25DD177 pKa = 3.76GLSITFVYY185 pKa = 10.46DD186 pKa = 3.48EE187 pKa = 5.15PRR189 pKa = 11.84SDD191 pKa = 3.29WEE193 pKa = 3.98ISVQPSPIAAHH204 pKa = 6.68ALAAVALGTEE214 pKa = 4.43DD215 pKa = 4.7AEE217 pKa = 4.36EE218 pKa = 4.48GKK220 pKa = 10.21QRR222 pKa = 11.84VVDD225 pKa = 4.01AFTNNVTEE233 pKa = 4.86DD234 pKa = 3.62LSALAKK240 pKa = 10.11SWNEE244 pKa = 3.67DD245 pKa = 3.53FNFTSLPEE253 pKa = 4.26DD254 pKa = 3.45EE255 pKa = 5.3NLYY258 pKa = 10.57LSSGPYY264 pKa = 10.56VMTDD268 pKa = 3.32YY269 pKa = 11.61VEE271 pKa = 4.1NQYY274 pKa = 9.01LTLSLRR280 pKa = 11.84EE281 pKa = 4.23DD282 pKa = 4.4YY283 pKa = 11.06SWGPMPSVDD292 pKa = 4.52EE293 pKa = 3.67ITIRR297 pKa = 11.84YY298 pKa = 9.48SEE300 pKa = 5.07DD301 pKa = 3.03PLASVTALQNGEE313 pKa = 3.81VDD315 pKa = 3.41IIEE318 pKa = 4.5PQASVDD324 pKa = 3.91VIEE327 pKa = 4.21TLEE330 pKa = 4.58GIEE333 pKa = 4.04GVEE336 pKa = 4.24YY337 pKa = 8.19TTAPTGTYY345 pKa = 8.83EE346 pKa = 4.54HH347 pKa = 7.15VDD349 pKa = 4.1LIFNNGGPFDD359 pKa = 4.17PATYY363 pKa = 10.55GGDD366 pKa = 3.42AAKK369 pKa = 10.53AQAVRR374 pKa = 11.84QAFLLSVPRR383 pKa = 11.84DD384 pKa = 3.56EE385 pKa = 6.1IIEE388 pKa = 4.32KK389 pKa = 10.13LVQPLQPDD397 pKa = 3.87AEE399 pKa = 4.34PRR401 pKa = 11.84EE402 pKa = 4.4SHH404 pKa = 5.71TVTSEE409 pKa = 3.51APAYY413 pKa = 10.39DD414 pKa = 3.97DD415 pKa = 4.21VVAGNGSEE423 pKa = 4.75FYY425 pKa = 9.71RR426 pKa = 11.84TSDD429 pKa = 3.49PAKK432 pKa = 10.19AAQLLADD439 pKa = 4.81AGVATPIDD447 pKa = 3.52VRR449 pKa = 11.84FLYY452 pKa = 10.46GASNVRR458 pKa = 11.84RR459 pKa = 11.84ANTFEE464 pKa = 4.92LIKK467 pKa = 10.92ASAAEE472 pKa = 3.95AGFNVIDD479 pKa = 4.79EE480 pKa = 5.18GDD482 pKa = 4.17DD483 pKa = 3.38NWGSRR488 pKa = 11.84LSDD491 pKa = 3.27TASYY495 pKa = 10.52DD496 pKa = 3.06ASLFGWQSTNTYY508 pKa = 10.53ALNSEE513 pKa = 4.56ANFVTGGLNNFAGMSDD529 pKa = 3.69PDD531 pKa = 3.07IDD533 pKa = 3.03AWYY536 pKa = 10.88AEE538 pKa = 4.49MSTNTDD544 pKa = 3.39PEE546 pKa = 4.32QEE548 pKa = 4.23TEE550 pKa = 4.0LTILIEE556 pKa = 4.06EE557 pKa = 4.49RR558 pKa = 11.84LNEE561 pKa = 4.31LGFSLPLYY569 pKa = 7.37QHH571 pKa = 7.06PGVMAYY577 pKa = 9.95RR578 pKa = 11.84DD579 pKa = 3.53SLQNVEE585 pKa = 5.0TIALSPTVFWNFWEE599 pKa = 4.38WEE601 pKa = 4.32VGGDD605 pKa = 3.72TADD608 pKa = 3.69EE609 pKa = 4.22EE610 pKa = 4.64
MM1 pKa = 7.44KK2 pKa = 9.61MRR4 pKa = 11.84RR5 pKa = 11.84IGAAGAIVATGALALSACSSTPSTTTEE32 pKa = 3.61IDD34 pKa = 3.41EE35 pKa = 4.35EE36 pKa = 4.61TSLSIGWNQPFYY48 pKa = 10.99SQNNLTSVGNATTNSNVIYY67 pKa = 9.01LTNAAFNYY75 pKa = 9.87YY76 pKa = 10.68DD77 pKa = 4.31NNLEE81 pKa = 4.09LQQFSDD87 pKa = 3.51FGTYY91 pKa = 7.97EE92 pKa = 4.08VEE94 pKa = 4.61SEE96 pKa = 4.46DD97 pKa = 4.36PLTVTYY103 pKa = 9.05TINEE107 pKa = 4.16GATWSDD113 pKa = 3.52GTPVDD118 pKa = 3.54AVDD121 pKa = 3.75VFLYY125 pKa = 9.92WGAQNDD131 pKa = 4.03SFDD134 pKa = 3.95NVAPEE139 pKa = 3.8YY140 pKa = 10.76DD141 pKa = 3.58EE142 pKa = 5.14EE143 pKa = 5.35GNITNQDD150 pKa = 3.7AIDD153 pKa = 3.75AGVYY157 pKa = 10.11FDD159 pKa = 4.39SSSVAMNLISEE170 pKa = 4.83PPTISDD176 pKa = 4.25DD177 pKa = 3.76GLSITFVYY185 pKa = 10.46DD186 pKa = 3.48EE187 pKa = 5.15PRR189 pKa = 11.84SDD191 pKa = 3.29WEE193 pKa = 3.98ISVQPSPIAAHH204 pKa = 6.68ALAAVALGTEE214 pKa = 4.43DD215 pKa = 4.7AEE217 pKa = 4.36EE218 pKa = 4.48GKK220 pKa = 10.21QRR222 pKa = 11.84VVDD225 pKa = 4.01AFTNNVTEE233 pKa = 4.86DD234 pKa = 3.62LSALAKK240 pKa = 10.11SWNEE244 pKa = 3.67DD245 pKa = 3.53FNFTSLPEE253 pKa = 4.26DD254 pKa = 3.45EE255 pKa = 5.3NLYY258 pKa = 10.57LSSGPYY264 pKa = 10.56VMTDD268 pKa = 3.32YY269 pKa = 11.61VEE271 pKa = 4.1NQYY274 pKa = 9.01LTLSLRR280 pKa = 11.84EE281 pKa = 4.23DD282 pKa = 4.4YY283 pKa = 11.06SWGPMPSVDD292 pKa = 4.52EE293 pKa = 3.67ITIRR297 pKa = 11.84YY298 pKa = 9.48SEE300 pKa = 5.07DD301 pKa = 3.03PLASVTALQNGEE313 pKa = 3.81VDD315 pKa = 3.41IIEE318 pKa = 4.5PQASVDD324 pKa = 3.91VIEE327 pKa = 4.21TLEE330 pKa = 4.58GIEE333 pKa = 4.04GVEE336 pKa = 4.24YY337 pKa = 8.19TTAPTGTYY345 pKa = 8.83EE346 pKa = 4.54HH347 pKa = 7.15VDD349 pKa = 4.1LIFNNGGPFDD359 pKa = 4.17PATYY363 pKa = 10.55GGDD366 pKa = 3.42AAKK369 pKa = 10.53AQAVRR374 pKa = 11.84QAFLLSVPRR383 pKa = 11.84DD384 pKa = 3.56EE385 pKa = 6.1IIEE388 pKa = 4.32KK389 pKa = 10.13LVQPLQPDD397 pKa = 3.87AEE399 pKa = 4.34PRR401 pKa = 11.84EE402 pKa = 4.4SHH404 pKa = 5.71TVTSEE409 pKa = 3.51APAYY413 pKa = 10.39DD414 pKa = 3.97DD415 pKa = 4.21VVAGNGSEE423 pKa = 4.75FYY425 pKa = 9.71RR426 pKa = 11.84TSDD429 pKa = 3.49PAKK432 pKa = 10.19AAQLLADD439 pKa = 4.81AGVATPIDD447 pKa = 3.52VRR449 pKa = 11.84FLYY452 pKa = 10.46GASNVRR458 pKa = 11.84RR459 pKa = 11.84ANTFEE464 pKa = 4.92LIKK467 pKa = 10.92ASAAEE472 pKa = 3.95AGFNVIDD479 pKa = 4.79EE480 pKa = 5.18GDD482 pKa = 4.17DD483 pKa = 3.38NWGSRR488 pKa = 11.84LSDD491 pKa = 3.27TASYY495 pKa = 10.52DD496 pKa = 3.06ASLFGWQSTNTYY508 pKa = 10.53ALNSEE513 pKa = 4.56ANFVTGGLNNFAGMSDD529 pKa = 3.69PDD531 pKa = 3.07IDD533 pKa = 3.03AWYY536 pKa = 10.88AEE538 pKa = 4.49MSTNTDD544 pKa = 3.39PEE546 pKa = 4.32QEE548 pKa = 4.23TEE550 pKa = 4.0LTILIEE556 pKa = 4.06EE557 pKa = 4.49RR558 pKa = 11.84LNEE561 pKa = 4.31LGFSLPLYY569 pKa = 7.37QHH571 pKa = 7.06PGVMAYY577 pKa = 9.95RR578 pKa = 11.84DD579 pKa = 3.53SLQNVEE585 pKa = 5.0TIALSPTVFWNFWEE599 pKa = 4.38WEE601 pKa = 4.32VGGDD605 pKa = 3.72TADD608 pKa = 3.69EE609 pKa = 4.22EE610 pKa = 4.64
Molecular weight: 66.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0BXG6|A0A0A0BXG6_9CELL Ribonucleoside-triphosphate reductase OS=Cellulomonas carbonis T26 OX=947969 GN=N868_07230 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.97GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.97GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1114276 |
35 |
1968 |
327.1 |
34.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.316 ± 0.055 | 0.593 ± 0.01 |
6.591 ± 0.035 | 5.6 ± 0.044 |
2.42 ± 0.026 | 9.481 ± 0.039 |
2.186 ± 0.02 | 2.556 ± 0.032 |
1.244 ± 0.027 | 10.251 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.608 ± 0.016 | 1.379 ± 0.022 |
5.955 ± 0.036 | 2.425 ± 0.021 |
8.307 ± 0.045 | 4.722 ± 0.024 |
6.319 ± 0.038 | 10.805 ± 0.051 |
1.497 ± 0.02 | 1.741 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
