
Bat polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
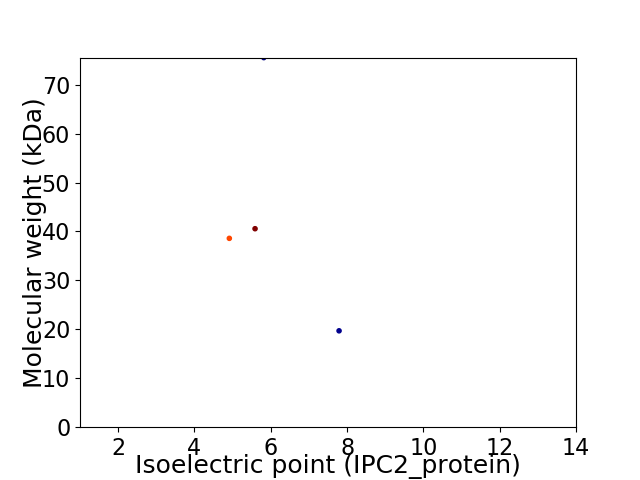
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3MCH7|A0A0D3MCH7_9POLY Capsid protein VP1 OS=Bat polyomavirus OX=1221637 PE=3 SV=1
MM1 pKa = 7.32GAFLAVLAEE10 pKa = 4.12VFEE13 pKa = 4.94LSTVTGLSVEE23 pKa = 4.57TILSGEE29 pKa = 4.19AFTTAEE35 pKa = 4.49LLQSHH40 pKa = 7.16IANLVTFGGLTEE52 pKa = 4.58AEE54 pKa = 3.95ALAAAEE60 pKa = 4.44VSAEE64 pKa = 4.14AYY66 pKa = 9.4SALSTLSEE74 pKa = 4.26TFPQAFATLAATEE87 pKa = 4.17FATTGTLTVGAAIAAALYY105 pKa = 9.05PYY107 pKa = 10.11YY108 pKa = 10.41YY109 pKa = 9.96DD110 pKa = 3.53YY111 pKa = 11.49SVPISNLNRR120 pKa = 11.84SMALQQWIPDD130 pKa = 3.67YY131 pKa = 11.47DD132 pKa = 4.39LDD134 pKa = 4.33FPGVRR139 pKa = 11.84PLVRR143 pKa = 11.84FLNYY147 pKa = 9.28IDD149 pKa = 5.05PYY151 pKa = 9.41TWASDD156 pKa = 3.75LYY158 pKa = 10.54HH159 pKa = 7.21SIGRR163 pKa = 11.84YY164 pKa = 7.36FWEE167 pKa = 3.99TAQRR171 pKa = 11.84IGQNVIEE178 pKa = 4.87EE179 pKa = 4.45EE180 pKa = 4.26LTDD183 pKa = 3.73LTRR186 pKa = 11.84DD187 pKa = 3.3LAVRR191 pKa = 11.84SVTSISEE198 pKa = 4.22MIAHH202 pKa = 6.01YY203 pKa = 10.34FEE205 pKa = 4.03NARR208 pKa = 11.84WAVSLLPRR216 pKa = 11.84SIYY219 pKa = 10.73NSLEE223 pKa = 3.92TYY225 pKa = 8.7YY226 pKa = 10.86TEE228 pKa = 4.99LPGINPIQARR238 pKa = 11.84QLYY241 pKa = 8.75RR242 pKa = 11.84KK243 pKa = 10.03LGEE246 pKa = 4.25TPIHH250 pKa = 7.02RR251 pKa = 11.84GRR253 pKa = 11.84GPGGPARR260 pKa = 11.84VRR262 pKa = 11.84RR263 pKa = 11.84SPYY266 pKa = 9.93LGRR269 pKa = 11.84GPGGPATIFGNIQGGAVPQQFSAQYY294 pKa = 9.04VEE296 pKa = 5.64RR297 pKa = 11.84YY298 pKa = 7.17PPPGGADD305 pKa = 2.91QRR307 pKa = 11.84TAADD311 pKa = 3.13WMLPLILGLYY321 pKa = 10.37GDD323 pKa = 5.15ISPSWRR329 pKa = 11.84RR330 pKa = 11.84TLEE333 pKa = 4.14DD334 pKa = 3.88LEE336 pKa = 4.44EE337 pKa = 4.75EE338 pKa = 4.1EE339 pKa = 6.42DD340 pKa = 4.22GPQKK344 pKa = 10.53KK345 pKa = 9.6KK346 pKa = 10.29PRR348 pKa = 11.84RR349 pKa = 11.84AA350 pKa = 3.22
MM1 pKa = 7.32GAFLAVLAEE10 pKa = 4.12VFEE13 pKa = 4.94LSTVTGLSVEE23 pKa = 4.57TILSGEE29 pKa = 4.19AFTTAEE35 pKa = 4.49LLQSHH40 pKa = 7.16IANLVTFGGLTEE52 pKa = 4.58AEE54 pKa = 3.95ALAAAEE60 pKa = 4.44VSAEE64 pKa = 4.14AYY66 pKa = 9.4SALSTLSEE74 pKa = 4.26TFPQAFATLAATEE87 pKa = 4.17FATTGTLTVGAAIAAALYY105 pKa = 9.05PYY107 pKa = 10.11YY108 pKa = 10.41YY109 pKa = 9.96DD110 pKa = 3.53YY111 pKa = 11.49SVPISNLNRR120 pKa = 11.84SMALQQWIPDD130 pKa = 3.67YY131 pKa = 11.47DD132 pKa = 4.39LDD134 pKa = 4.33FPGVRR139 pKa = 11.84PLVRR143 pKa = 11.84FLNYY147 pKa = 9.28IDD149 pKa = 5.05PYY151 pKa = 9.41TWASDD156 pKa = 3.75LYY158 pKa = 10.54HH159 pKa = 7.21SIGRR163 pKa = 11.84YY164 pKa = 7.36FWEE167 pKa = 3.99TAQRR171 pKa = 11.84IGQNVIEE178 pKa = 4.87EE179 pKa = 4.45EE180 pKa = 4.26LTDD183 pKa = 3.73LTRR186 pKa = 11.84DD187 pKa = 3.3LAVRR191 pKa = 11.84SVTSISEE198 pKa = 4.22MIAHH202 pKa = 6.01YY203 pKa = 10.34FEE205 pKa = 4.03NARR208 pKa = 11.84WAVSLLPRR216 pKa = 11.84SIYY219 pKa = 10.73NSLEE223 pKa = 3.92TYY225 pKa = 8.7YY226 pKa = 10.86TEE228 pKa = 4.99LPGINPIQARR238 pKa = 11.84QLYY241 pKa = 8.75RR242 pKa = 11.84KK243 pKa = 10.03LGEE246 pKa = 4.25TPIHH250 pKa = 7.02RR251 pKa = 11.84GRR253 pKa = 11.84GPGGPARR260 pKa = 11.84VRR262 pKa = 11.84RR263 pKa = 11.84SPYY266 pKa = 9.93LGRR269 pKa = 11.84GPGGPATIFGNIQGGAVPQQFSAQYY294 pKa = 9.04VEE296 pKa = 5.64RR297 pKa = 11.84YY298 pKa = 7.17PPPGGADD305 pKa = 2.91QRR307 pKa = 11.84TAADD311 pKa = 3.13WMLPLILGLYY321 pKa = 10.37GDD323 pKa = 5.15ISPSWRR329 pKa = 11.84RR330 pKa = 11.84TLEE333 pKa = 4.14DD334 pKa = 3.88LEE336 pKa = 4.44EE337 pKa = 4.75EE338 pKa = 4.1EE339 pKa = 6.42DD340 pKa = 4.22GPQKK344 pKa = 10.53KK345 pKa = 9.6KK346 pKa = 10.29PRR348 pKa = 11.84RR349 pKa = 11.84AA350 pKa = 3.22
Molecular weight: 38.59 kDa
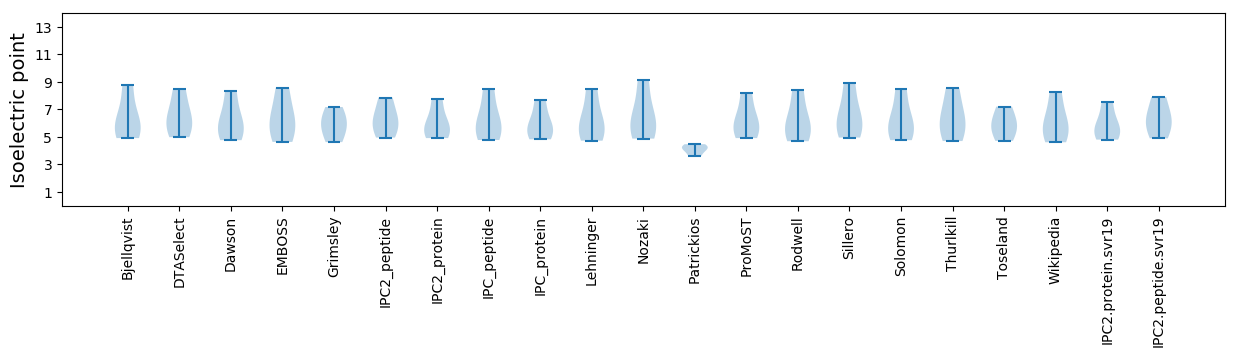
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3MDQ1|A0A0D3MDQ1_9POLY Large T antigen OS=Bat polyomavirus OX=1221637 PE=4 SV=1
MM1 pKa = 7.85DD2 pKa = 3.57HH3 pKa = 6.67TLTRR7 pKa = 11.84EE8 pKa = 3.46EE9 pKa = 4.37SLRR12 pKa = 11.84LMEE15 pKa = 5.3LLSLPMEE22 pKa = 4.35QYY24 pKa = 11.84GNFPLMRR31 pKa = 11.84KK32 pKa = 9.48AFLKK36 pKa = 10.02KK37 pKa = 10.34CKK39 pKa = 9.55ILHH42 pKa = 6.42PDD44 pKa = 2.78KK45 pKa = 11.36GGNQEE50 pKa = 4.08LAKK53 pKa = 10.45EE54 pKa = 4.36LISLYY59 pKa = 10.52KK60 pKa = 10.27RR61 pKa = 11.84LEE63 pKa = 4.09EE64 pKa = 4.17MLPTLNPEE72 pKa = 4.14EE73 pKa = 4.55SFSTSQVHH81 pKa = 5.85EE82 pKa = 4.36SSDD85 pKa = 3.45LFIWMDD91 pKa = 3.73DD92 pKa = 3.25WPKK95 pKa = 11.02CKK97 pKa = 9.86IGANPTCRR105 pKa = 11.84CLFCLLQRR113 pKa = 11.84NHH115 pKa = 6.59RR116 pKa = 11.84KK117 pKa = 9.37RR118 pKa = 11.84RR119 pKa = 11.84TPWPKK124 pKa = 9.25VWGDD128 pKa = 3.59CFCFRR133 pKa = 11.84CYY135 pKa = 9.21ITWFGLEE142 pKa = 4.03NSWMVFLSWKK152 pKa = 9.59EE153 pKa = 3.9IVGKK157 pKa = 8.41TPYY160 pKa = 9.94RR161 pKa = 11.84VLNII165 pKa = 3.89
MM1 pKa = 7.85DD2 pKa = 3.57HH3 pKa = 6.67TLTRR7 pKa = 11.84EE8 pKa = 3.46EE9 pKa = 4.37SLRR12 pKa = 11.84LMEE15 pKa = 5.3LLSLPMEE22 pKa = 4.35QYY24 pKa = 11.84GNFPLMRR31 pKa = 11.84KK32 pKa = 9.48AFLKK36 pKa = 10.02KK37 pKa = 10.34CKK39 pKa = 9.55ILHH42 pKa = 6.42PDD44 pKa = 2.78KK45 pKa = 11.36GGNQEE50 pKa = 4.08LAKK53 pKa = 10.45EE54 pKa = 4.36LISLYY59 pKa = 10.52KK60 pKa = 10.27RR61 pKa = 11.84LEE63 pKa = 4.09EE64 pKa = 4.17MLPTLNPEE72 pKa = 4.14EE73 pKa = 4.55SFSTSQVHH81 pKa = 5.85EE82 pKa = 4.36SSDD85 pKa = 3.45LFIWMDD91 pKa = 3.73DD92 pKa = 3.25WPKK95 pKa = 11.02CKK97 pKa = 9.86IGANPTCRR105 pKa = 11.84CLFCLLQRR113 pKa = 11.84NHH115 pKa = 6.59RR116 pKa = 11.84KK117 pKa = 9.37RR118 pKa = 11.84RR119 pKa = 11.84TPWPKK124 pKa = 9.25VWGDD128 pKa = 3.59CFCFRR133 pKa = 11.84CYY135 pKa = 9.21ITWFGLEE142 pKa = 4.03NSWMVFLSWKK152 pKa = 9.59EE153 pKa = 3.9IVGKK157 pKa = 8.41TPYY160 pKa = 9.94RR161 pKa = 11.84VLNII165 pKa = 3.89
Molecular weight: 19.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1540 |
165 |
657 |
385.0 |
43.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.558 ± 1.349 | 2.143 ± 0.624 |
4.935 ± 0.544 | 6.948 ± 0.36 |
5.26 ± 0.923 | 6.039 ± 0.909 |
1.688 ± 0.26 | 5.0 ± 0.193 |
5.714 ± 1.386 | 10.455 ± 0.593 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.468 ± 0.402 | 4.416 ± 0.596 |
6.299 ± 0.623 | 4.675 ± 0.493 |
5.0 ± 0.656 | 5.779 ± 0.31 |
6.169 ± 0.381 | 5.519 ± 0.682 |
1.558 ± 0.424 | 3.377 ± 0.723 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
