
Jannaschia seohaensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Jannaschia
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
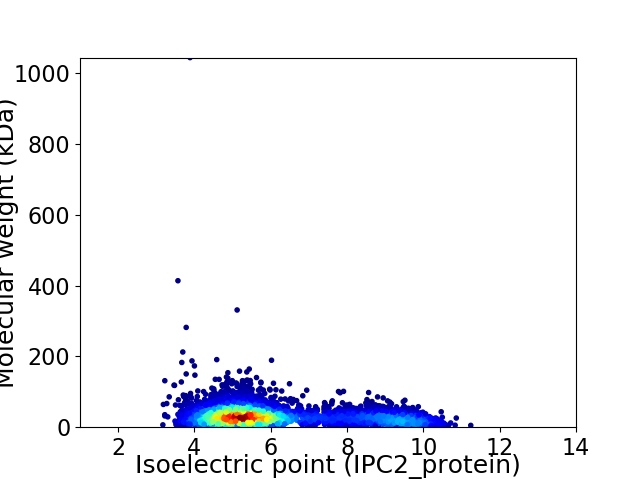
Virtual 2D-PAGE plot for 4631 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Y9B033|A0A2Y9B033_9RHOB Uncharacterized protein OS=Jannaschia seohaensis OX=475081 GN=BCF38_10896 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 10.32KK3 pKa = 9.94LASSTAIAVLLSSAAYY19 pKa = 10.34ADD21 pKa = 3.52GHH23 pKa = 5.67TAAFTPGEE31 pKa = 4.59GPFDD35 pKa = 3.63WDD37 pKa = 3.57SYY39 pKa = 10.48NAFAEE44 pKa = 4.37ATDD47 pKa = 3.96LTGQTVTVTGPWTGDD62 pKa = 2.85EE63 pKa = 4.06AAKK66 pKa = 10.35FDD68 pKa = 3.45IVMAFFADD76 pKa = 3.41ATGAEE81 pKa = 4.46VNYY84 pKa = 10.53SGSDD88 pKa = 3.18SFEE91 pKa = 3.7QDD93 pKa = 2.49IVISSRR99 pKa = 11.84AGSAPNLAVFPQPGLAADD117 pKa = 3.95MASQGLLTPLPDD129 pKa = 3.7GTADD133 pKa = 3.04WVAEE137 pKa = 4.0NFAAGQSWVDD147 pKa = 3.33LGTYY151 pKa = 10.06AGEE154 pKa = 4.55DD155 pKa = 4.12GEE157 pKa = 5.35DD158 pKa = 3.42DD159 pKa = 4.38LYY161 pKa = 11.88GMFYY165 pKa = 10.51RR166 pKa = 11.84VDD168 pKa = 3.71LKK170 pKa = 11.43SLVWYY175 pKa = 10.44SPAAFDD181 pKa = 4.69EE182 pKa = 4.24MGYY185 pKa = 10.38DD186 pKa = 2.95IPTTMEE192 pKa = 3.96EE193 pKa = 4.35LKK195 pKa = 10.74EE196 pKa = 3.93LTDD199 pKa = 3.67QIVADD204 pKa = 5.74GEE206 pKa = 4.8TPWCIGLGSGAATGWPATDD225 pKa = 3.12WVEE228 pKa = 5.29DD229 pKa = 3.55MMLRR233 pKa = 11.84TQPPSVYY240 pKa = 10.17DD241 pKa = 3.08AWVSNEE247 pKa = 4.22LPFDD251 pKa = 3.96APEE254 pKa = 3.85VVAAIEE260 pKa = 4.05EE261 pKa = 4.19FGYY264 pKa = 10.0FARR267 pKa = 11.84NDD269 pKa = 3.81DD270 pKa = 4.09YY271 pKa = 12.1VSGGSQAVASTDD283 pKa = 3.98FRR285 pKa = 11.84DD286 pKa = 3.68SPTGLFAIPPACYY299 pKa = 8.65MHH301 pKa = 6.78RR302 pKa = 11.84QASFIPAFFPEE313 pKa = 4.61GTVVGEE319 pKa = 4.55DD320 pKa = 2.7VDD322 pKa = 5.26FFYY325 pKa = 11.13FPAYY329 pKa = 10.05ADD331 pKa = 3.98KK332 pKa = 11.33DD333 pKa = 3.54LGAPVLGAGTLFAITNPSEE352 pKa = 3.87GAEE355 pKa = 3.92AMIEE359 pKa = 4.06FLKK362 pKa = 11.07LPIAHH367 pKa = 6.73EE368 pKa = 3.96LWMAQGGFLTAHH380 pKa = 6.9AGVNTDD386 pKa = 4.15AYY388 pKa = 11.27SEE390 pKa = 4.36DD391 pKa = 3.63SLRR394 pKa = 11.84AQGQILLDD402 pKa = 3.38ATTFRR407 pKa = 11.84FDD409 pKa = 5.22ASDD412 pKa = 3.81LMPGEE417 pKa = 4.37IGAGAFWTGMVDD429 pKa = 3.67YY430 pKa = 9.84VTGASAEE437 pKa = 4.09EE438 pKa = 4.18VAAGVQEE445 pKa = 4.55RR446 pKa = 11.84WQSLNN451 pKa = 3.22
MM1 pKa = 7.58KK2 pKa = 10.32KK3 pKa = 9.94LASSTAIAVLLSSAAYY19 pKa = 10.34ADD21 pKa = 3.52GHH23 pKa = 5.67TAAFTPGEE31 pKa = 4.59GPFDD35 pKa = 3.63WDD37 pKa = 3.57SYY39 pKa = 10.48NAFAEE44 pKa = 4.37ATDD47 pKa = 3.96LTGQTVTVTGPWTGDD62 pKa = 2.85EE63 pKa = 4.06AAKK66 pKa = 10.35FDD68 pKa = 3.45IVMAFFADD76 pKa = 3.41ATGAEE81 pKa = 4.46VNYY84 pKa = 10.53SGSDD88 pKa = 3.18SFEE91 pKa = 3.7QDD93 pKa = 2.49IVISSRR99 pKa = 11.84AGSAPNLAVFPQPGLAADD117 pKa = 3.95MASQGLLTPLPDD129 pKa = 3.7GTADD133 pKa = 3.04WVAEE137 pKa = 4.0NFAAGQSWVDD147 pKa = 3.33LGTYY151 pKa = 10.06AGEE154 pKa = 4.55DD155 pKa = 4.12GEE157 pKa = 5.35DD158 pKa = 3.42DD159 pKa = 4.38LYY161 pKa = 11.88GMFYY165 pKa = 10.51RR166 pKa = 11.84VDD168 pKa = 3.71LKK170 pKa = 11.43SLVWYY175 pKa = 10.44SPAAFDD181 pKa = 4.69EE182 pKa = 4.24MGYY185 pKa = 10.38DD186 pKa = 2.95IPTTMEE192 pKa = 3.96EE193 pKa = 4.35LKK195 pKa = 10.74EE196 pKa = 3.93LTDD199 pKa = 3.67QIVADD204 pKa = 5.74GEE206 pKa = 4.8TPWCIGLGSGAATGWPATDD225 pKa = 3.12WVEE228 pKa = 5.29DD229 pKa = 3.55MMLRR233 pKa = 11.84TQPPSVYY240 pKa = 10.17DD241 pKa = 3.08AWVSNEE247 pKa = 4.22LPFDD251 pKa = 3.96APEE254 pKa = 3.85VVAAIEE260 pKa = 4.05EE261 pKa = 4.19FGYY264 pKa = 10.0FARR267 pKa = 11.84NDD269 pKa = 3.81DD270 pKa = 4.09YY271 pKa = 12.1VSGGSQAVASTDD283 pKa = 3.98FRR285 pKa = 11.84DD286 pKa = 3.68SPTGLFAIPPACYY299 pKa = 8.65MHH301 pKa = 6.78RR302 pKa = 11.84QASFIPAFFPEE313 pKa = 4.61GTVVGEE319 pKa = 4.55DD320 pKa = 2.7VDD322 pKa = 5.26FFYY325 pKa = 11.13FPAYY329 pKa = 10.05ADD331 pKa = 3.98KK332 pKa = 11.33DD333 pKa = 3.54LGAPVLGAGTLFAITNPSEE352 pKa = 3.87GAEE355 pKa = 3.92AMIEE359 pKa = 4.06FLKK362 pKa = 11.07LPIAHH367 pKa = 6.73EE368 pKa = 3.96LWMAQGGFLTAHH380 pKa = 6.9AGVNTDD386 pKa = 4.15AYY388 pKa = 11.27SEE390 pKa = 4.36DD391 pKa = 3.63SLRR394 pKa = 11.84AQGQILLDD402 pKa = 3.38ATTFRR407 pKa = 11.84FDD409 pKa = 5.22ASDD412 pKa = 3.81LMPGEE417 pKa = 4.37IGAGAFWTGMVDD429 pKa = 3.67YY430 pKa = 9.84VTGASAEE437 pKa = 4.09EE438 pKa = 4.18VAAGVQEE445 pKa = 4.55RR446 pKa = 11.84WQSLNN451 pKa = 3.22
Molecular weight: 48.11 kDa
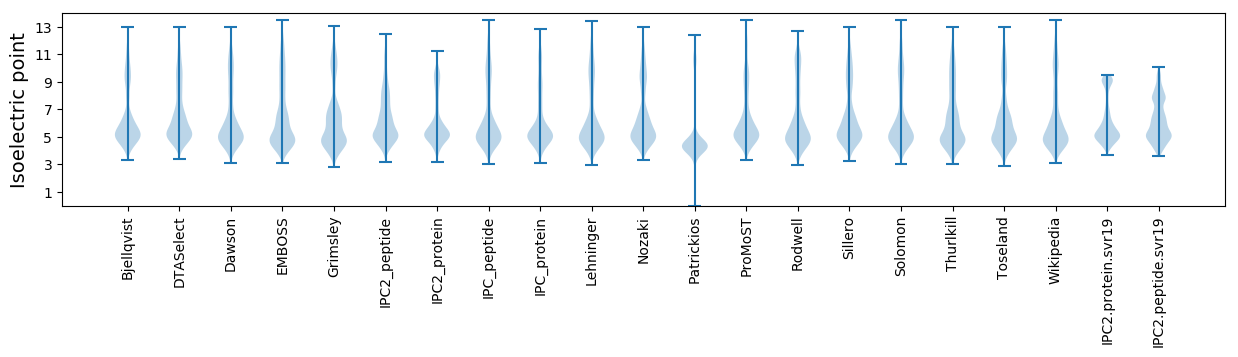
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Y9AB67|A0A2Y9AB67_9RHOB Uncharacterized protein OS=Jannaschia seohaensis OX=475081 GN=BCF38_102252 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1425063 |
28 |
10108 |
307.7 |
33.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.525 ± 0.064 | 0.842 ± 0.012 |
6.208 ± 0.049 | 6.019 ± 0.034 |
3.464 ± 0.024 | 9.173 ± 0.054 |
1.959 ± 0.018 | 4.643 ± 0.03 |
2.267 ± 0.031 | 10.35 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.024 | 2.054 ± 0.024 |
5.593 ± 0.034 | 2.747 ± 0.018 |
7.556 ± 0.052 | 4.739 ± 0.027 |
5.513 ± 0.033 | 7.379 ± 0.029 |
1.477 ± 0.021 | 1.948 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
