
Rhizoctonia solani dsRNA virus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
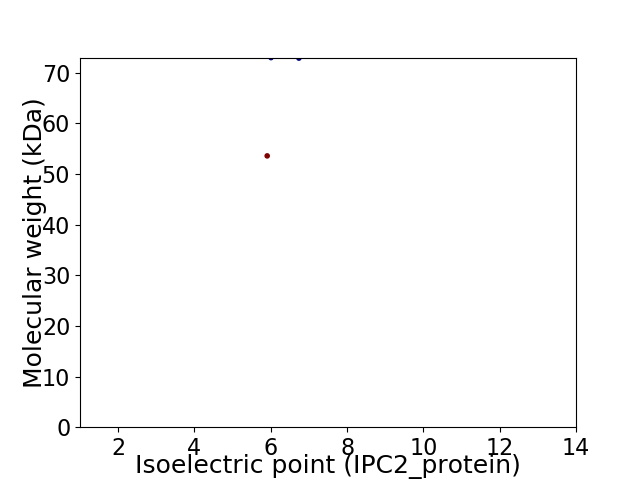
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D1BPE2|A0A2D1BPE2_9VIRU RNA-dependent RNA polymerase OS=Rhizoctonia solani dsRNA virus 4 OX=2045504 PE=4 SV=1
MM1 pKa = 7.57TDD3 pKa = 4.33KK4 pKa = 10.7ISQSTVGAIAAEE16 pKa = 4.27PSPPAGPAPATRR28 pKa = 11.84AEE30 pKa = 4.61PNPPPAPTATSKK42 pKa = 10.96PKK44 pKa = 10.73GKK46 pKa = 9.86LRR48 pKa = 11.84PASNFTPTSHH58 pKa = 7.06NGIAPMLQVVSNRR71 pKa = 11.84RR72 pKa = 11.84SHH74 pKa = 6.97IINRR78 pKa = 11.84AEE80 pKa = 3.86PSGLFPSAINYY91 pKa = 7.15FQAVSITDD99 pKa = 3.26QRR101 pKa = 11.84MSTTKK106 pKa = 10.51KK107 pKa = 10.04FLEE110 pKa = 4.74TSEE113 pKa = 4.04SWHH116 pKa = 6.21PVVSQYY122 pKa = 9.66YY123 pKa = 8.6ACFLFTVQVLRR134 pKa = 11.84TYY136 pKa = 10.41NVSKK140 pKa = 10.87IPDD143 pKa = 3.72TEE145 pKa = 4.14GLDD148 pKa = 3.31FLSYY152 pKa = 11.04VEE154 pKa = 4.41EE155 pKa = 4.17QLDD158 pKa = 3.81LTQIPVPGTWAEE170 pKa = 3.86YY171 pKa = 10.25LRR173 pKa = 11.84NITVVEE179 pKa = 3.93AHH181 pKa = 6.88HH182 pKa = 7.2DD183 pKa = 3.72SFGNIHH189 pKa = 6.65PVFPLLGGSPLTAGDD204 pKa = 3.75VFSFPQSHH212 pKa = 6.29RR213 pKa = 11.84RR214 pKa = 11.84ILPNCYY220 pKa = 9.57LALDD224 pKa = 3.54QLIRR228 pKa = 11.84YY229 pKa = 7.25AQTGANTNSPRR240 pKa = 11.84FINYY244 pKa = 10.12ANIFQLNPTTSDD256 pKa = 2.79EE257 pKa = 4.06AAQAKK262 pKa = 8.31LAPQVSSWSHH272 pKa = 4.42VSNSRR277 pKa = 11.84NDD279 pKa = 3.08VSLEE283 pKa = 3.72FWANAATLLPDD294 pKa = 3.91RR295 pKa = 11.84ADD297 pKa = 3.25SRR299 pKa = 11.84TGHH302 pKa = 6.64HH303 pKa = 6.65SGDD306 pKa = 3.44VTDD309 pKa = 5.8FFVFTGFRR317 pKa = 11.84NAAGTASHH325 pKa = 6.66SFFDD329 pKa = 5.51VILQDD334 pKa = 3.34MSSYY338 pKa = 10.18CQFVKK343 pKa = 10.91GSVPLSTIQSIGIGATIPVMYY364 pKa = 9.03VQPSTSVRR372 pKa = 11.84NYY374 pKa = 8.8FFPSLADD381 pKa = 3.78LRR383 pKa = 11.84DD384 pKa = 3.31NTNYY388 pKa = 9.57HH389 pKa = 6.55RR390 pKa = 11.84SGHH393 pKa = 5.87RR394 pKa = 11.84LFPTTITYY402 pKa = 11.01DD403 pKa = 3.39MRR405 pKa = 11.84HH406 pKa = 5.93GDD408 pKa = 3.48LTLEE412 pKa = 4.68EE413 pKa = 4.4IAEE416 pKa = 4.23QYY418 pKa = 11.57AKK420 pKa = 9.42LTCLNTSFRR429 pKa = 11.84DD430 pKa = 3.58LPTQNGWNQPDD441 pKa = 3.73EE442 pKa = 4.5TTVRR446 pKa = 11.84DD447 pKa = 4.11GPFWAITEE455 pKa = 4.27YY456 pKa = 11.09KK457 pKa = 9.7VDD459 pKa = 3.64RR460 pKa = 11.84GVNPITGLLTNIPAYY475 pKa = 9.49YY476 pKa = 9.94HH477 pKa = 6.37SPNAVANKK485 pKa = 10.01
MM1 pKa = 7.57TDD3 pKa = 4.33KK4 pKa = 10.7ISQSTVGAIAAEE16 pKa = 4.27PSPPAGPAPATRR28 pKa = 11.84AEE30 pKa = 4.61PNPPPAPTATSKK42 pKa = 10.96PKK44 pKa = 10.73GKK46 pKa = 9.86LRR48 pKa = 11.84PASNFTPTSHH58 pKa = 7.06NGIAPMLQVVSNRR71 pKa = 11.84RR72 pKa = 11.84SHH74 pKa = 6.97IINRR78 pKa = 11.84AEE80 pKa = 3.86PSGLFPSAINYY91 pKa = 7.15FQAVSITDD99 pKa = 3.26QRR101 pKa = 11.84MSTTKK106 pKa = 10.51KK107 pKa = 10.04FLEE110 pKa = 4.74TSEE113 pKa = 4.04SWHH116 pKa = 6.21PVVSQYY122 pKa = 9.66YY123 pKa = 8.6ACFLFTVQVLRR134 pKa = 11.84TYY136 pKa = 10.41NVSKK140 pKa = 10.87IPDD143 pKa = 3.72TEE145 pKa = 4.14GLDD148 pKa = 3.31FLSYY152 pKa = 11.04VEE154 pKa = 4.41EE155 pKa = 4.17QLDD158 pKa = 3.81LTQIPVPGTWAEE170 pKa = 3.86YY171 pKa = 10.25LRR173 pKa = 11.84NITVVEE179 pKa = 3.93AHH181 pKa = 6.88HH182 pKa = 7.2DD183 pKa = 3.72SFGNIHH189 pKa = 6.65PVFPLLGGSPLTAGDD204 pKa = 3.75VFSFPQSHH212 pKa = 6.29RR213 pKa = 11.84RR214 pKa = 11.84ILPNCYY220 pKa = 9.57LALDD224 pKa = 3.54QLIRR228 pKa = 11.84YY229 pKa = 7.25AQTGANTNSPRR240 pKa = 11.84FINYY244 pKa = 10.12ANIFQLNPTTSDD256 pKa = 2.79EE257 pKa = 4.06AAQAKK262 pKa = 8.31LAPQVSSWSHH272 pKa = 4.42VSNSRR277 pKa = 11.84NDD279 pKa = 3.08VSLEE283 pKa = 3.72FWANAATLLPDD294 pKa = 3.91RR295 pKa = 11.84ADD297 pKa = 3.25SRR299 pKa = 11.84TGHH302 pKa = 6.64HH303 pKa = 6.65SGDD306 pKa = 3.44VTDD309 pKa = 5.8FFVFTGFRR317 pKa = 11.84NAAGTASHH325 pKa = 6.66SFFDD329 pKa = 5.51VILQDD334 pKa = 3.34MSSYY338 pKa = 10.18CQFVKK343 pKa = 10.91GSVPLSTIQSIGIGATIPVMYY364 pKa = 9.03VQPSTSVRR372 pKa = 11.84NYY374 pKa = 8.8FFPSLADD381 pKa = 3.78LRR383 pKa = 11.84DD384 pKa = 3.31NTNYY388 pKa = 9.57HH389 pKa = 6.55RR390 pKa = 11.84SGHH393 pKa = 5.87RR394 pKa = 11.84LFPTTITYY402 pKa = 11.01DD403 pKa = 3.39MRR405 pKa = 11.84HH406 pKa = 5.93GDD408 pKa = 3.48LTLEE412 pKa = 4.68EE413 pKa = 4.4IAEE416 pKa = 4.23QYY418 pKa = 11.57AKK420 pKa = 9.42LTCLNTSFRR429 pKa = 11.84DD430 pKa = 3.58LPTQNGWNQPDD441 pKa = 3.73EE442 pKa = 4.5TTVRR446 pKa = 11.84DD447 pKa = 4.11GPFWAITEE455 pKa = 4.27YY456 pKa = 11.09KK457 pKa = 9.7VDD459 pKa = 3.64RR460 pKa = 11.84GVNPITGLLTNIPAYY475 pKa = 9.49YY476 pKa = 9.94HH477 pKa = 6.37SPNAVANKK485 pKa = 10.01
Molecular weight: 53.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D1BPE2|A0A2D1BPE2_9VIRU RNA-dependent RNA polymerase OS=Rhizoctonia solani dsRNA virus 4 OX=2045504 PE=4 SV=1
MM1 pKa = 7.64EE2 pKa = 5.22YY3 pKa = 10.23INRR6 pKa = 11.84VLSYY10 pKa = 9.89IFKK13 pKa = 10.56PKK15 pKa = 10.01FEE17 pKa = 4.6EE18 pKa = 4.29YY19 pKa = 11.16NFIEE23 pKa = 4.12VGRR26 pKa = 11.84YY27 pKa = 8.25SRR29 pKa = 11.84PPHH32 pKa = 6.91VPTQDD37 pKa = 3.05TLSIEE42 pKa = 3.91KK43 pKa = 9.33HH44 pKa = 5.32QNVMMHH50 pKa = 6.05SMRR53 pKa = 11.84KK54 pKa = 9.05YY55 pKa = 9.07LYY57 pKa = 8.49PWEE60 pKa = 4.34IEE62 pKa = 3.97QIEE65 pKa = 4.88SYY67 pKa = 10.84RR68 pKa = 11.84RR69 pKa = 11.84TTTSEE74 pKa = 3.55SDD76 pKa = 2.95IEE78 pKa = 4.39RR79 pKa = 11.84DD80 pKa = 3.46FFTGNVEE87 pKa = 4.58HH88 pKa = 7.33FDD90 pKa = 3.95PPLDD94 pKa = 3.82EE95 pKa = 4.78NFEE98 pKa = 4.59LGLADD103 pKa = 3.68MLDD106 pKa = 3.75AFRR109 pKa = 11.84PPQKK113 pKa = 10.22CRR115 pKa = 11.84PCHH118 pKa = 5.9INDD121 pKa = 3.87IEE123 pKa = 4.02HH124 pKa = 7.15HH125 pKa = 6.91YY126 pKa = 9.06PFKK129 pKa = 10.09WQVNAEE135 pKa = 4.38APFSTDD141 pKa = 5.66DD142 pKa = 3.52YY143 pKa = 11.61FLDD146 pKa = 3.73NRR148 pKa = 11.84PKK150 pKa = 10.2ISEE153 pKa = 4.21LADD156 pKa = 3.28SKK158 pKa = 11.52YY159 pKa = 11.21AFLDD163 pKa = 3.04QRR165 pKa = 11.84YY166 pKa = 9.19VDD168 pKa = 4.44RR169 pKa = 11.84LLSKK173 pKa = 9.31QTSEE177 pKa = 4.23MNNPVPAKK185 pKa = 9.6FGPMRR190 pKa = 11.84HH191 pKa = 5.46TVFNWTRR198 pKa = 11.84RR199 pKa = 11.84WHH201 pKa = 6.44HH202 pKa = 5.72VIKK205 pKa = 10.89DD206 pKa = 3.34GFEE209 pKa = 3.86RR210 pKa = 11.84HH211 pKa = 5.95AHH213 pKa = 6.02LKK215 pKa = 11.12DD216 pKa = 3.29NDD218 pKa = 4.02PYY220 pKa = 10.96FQNKK224 pKa = 9.72YY225 pKa = 8.89IFPMLLHH232 pKa = 7.04AKK234 pKa = 8.27TAIVKK239 pKa = 10.28KK240 pKa = 10.44DD241 pKa = 3.95DD242 pKa = 4.07PNKK245 pKa = 9.8MRR247 pKa = 11.84TIWGCSKK254 pKa = 10.23PWIIADD260 pKa = 3.27AMLYY264 pKa = 9.31WEE266 pKa = 4.52YY267 pKa = 10.78IAWIKK272 pKa = 10.3LNPGLTPMLWGYY284 pKa = 7.79EE285 pKa = 4.05TMTGGWMRR293 pKa = 11.84LNAALFSSYY302 pKa = 11.1LNKK305 pKa = 10.48CYY307 pKa = 9.66LTLDD311 pKa = 3.49WSRR314 pKa = 11.84FDD316 pKa = 3.04KK317 pKa = 10.71RR318 pKa = 11.84AYY320 pKa = 9.91FSIIRR325 pKa = 11.84RR326 pKa = 11.84IMFGILTYY334 pKa = 10.95LDD336 pKa = 3.84FDD338 pKa = 3.65NGYY341 pKa = 9.48VPNVNYY347 pKa = 9.95PDD349 pKa = 3.94TKK351 pKa = 11.01SDD353 pKa = 3.09WSHH356 pKa = 6.63LKK358 pKa = 9.46SQRR361 pKa = 11.84ILRR364 pKa = 11.84LWLWTLDD371 pKa = 3.69NLFNAPIVLPDD382 pKa = 2.78GRR384 pKa = 11.84MYY386 pKa = 11.0KK387 pKa = 10.2RR388 pKa = 11.84LFAGIPSGLFITQLLDD404 pKa = 2.7SWYY407 pKa = 9.6NYY409 pKa = 9.02TMIATILRR417 pKa = 11.84AMGFNPRR424 pKa = 11.84QCIIKK429 pKa = 9.98VQGDD433 pKa = 3.6DD434 pKa = 3.9SIVRR438 pKa = 11.84LYY440 pKa = 11.33VLIPPAKK447 pKa = 10.39HH448 pKa = 6.2EE449 pKa = 4.37DD450 pKa = 3.29FLLRR454 pKa = 11.84MQEE457 pKa = 4.0LADD460 pKa = 4.14HH461 pKa = 6.04YY462 pKa = 9.39FKK464 pKa = 11.25SVISVDD470 pKa = 3.1KK471 pKa = 11.48SEE473 pKa = 5.01IGSSLNGRR481 pKa = 11.84EE482 pKa = 3.88VLSYY486 pKa = 10.61RR487 pKa = 11.84NKK489 pKa = 10.43NGFPFRR495 pKa = 11.84DD496 pKa = 4.15EE497 pKa = 4.08IKK499 pKa = 10.24MIAQFYY505 pKa = 7.08HH506 pKa = 5.15TKK508 pKa = 10.25ARR510 pKa = 11.84KK511 pKa = 7.28STPEE515 pKa = 3.28IAMAQAIGFAYY526 pKa = 10.15ASCGNHH532 pKa = 6.52DD533 pKa = 3.93RR534 pKa = 11.84VLSVLEE540 pKa = 4.37DD541 pKa = 3.54MYY543 pKa = 11.84DD544 pKa = 3.2HH545 pKa = 6.78YY546 pKa = 11.29LSQGYY551 pKa = 9.3SPNPAGLTMVFGNSPDD567 pKa = 3.64TEE569 pKa = 4.2RR570 pKa = 11.84PFIDD574 pKa = 3.4MDD576 pKa = 3.85HH577 pKa = 6.77FPSISEE583 pKa = 3.6IKK585 pKa = 10.34RR586 pKa = 11.84FLVCTDD592 pKa = 3.1YY593 pKa = 11.58RR594 pKa = 11.84NDD596 pKa = 3.39VQSRR600 pKa = 11.84RR601 pKa = 11.84TWPPDD606 pKa = 3.43YY607 pKa = 10.28FLHH610 pKa = 7.08PPACKK615 pKa = 10.12PEE617 pKa = 3.73
MM1 pKa = 7.64EE2 pKa = 5.22YY3 pKa = 10.23INRR6 pKa = 11.84VLSYY10 pKa = 9.89IFKK13 pKa = 10.56PKK15 pKa = 10.01FEE17 pKa = 4.6EE18 pKa = 4.29YY19 pKa = 11.16NFIEE23 pKa = 4.12VGRR26 pKa = 11.84YY27 pKa = 8.25SRR29 pKa = 11.84PPHH32 pKa = 6.91VPTQDD37 pKa = 3.05TLSIEE42 pKa = 3.91KK43 pKa = 9.33HH44 pKa = 5.32QNVMMHH50 pKa = 6.05SMRR53 pKa = 11.84KK54 pKa = 9.05YY55 pKa = 9.07LYY57 pKa = 8.49PWEE60 pKa = 4.34IEE62 pKa = 3.97QIEE65 pKa = 4.88SYY67 pKa = 10.84RR68 pKa = 11.84RR69 pKa = 11.84TTTSEE74 pKa = 3.55SDD76 pKa = 2.95IEE78 pKa = 4.39RR79 pKa = 11.84DD80 pKa = 3.46FFTGNVEE87 pKa = 4.58HH88 pKa = 7.33FDD90 pKa = 3.95PPLDD94 pKa = 3.82EE95 pKa = 4.78NFEE98 pKa = 4.59LGLADD103 pKa = 3.68MLDD106 pKa = 3.75AFRR109 pKa = 11.84PPQKK113 pKa = 10.22CRR115 pKa = 11.84PCHH118 pKa = 5.9INDD121 pKa = 3.87IEE123 pKa = 4.02HH124 pKa = 7.15HH125 pKa = 6.91YY126 pKa = 9.06PFKK129 pKa = 10.09WQVNAEE135 pKa = 4.38APFSTDD141 pKa = 5.66DD142 pKa = 3.52YY143 pKa = 11.61FLDD146 pKa = 3.73NRR148 pKa = 11.84PKK150 pKa = 10.2ISEE153 pKa = 4.21LADD156 pKa = 3.28SKK158 pKa = 11.52YY159 pKa = 11.21AFLDD163 pKa = 3.04QRR165 pKa = 11.84YY166 pKa = 9.19VDD168 pKa = 4.44RR169 pKa = 11.84LLSKK173 pKa = 9.31QTSEE177 pKa = 4.23MNNPVPAKK185 pKa = 9.6FGPMRR190 pKa = 11.84HH191 pKa = 5.46TVFNWTRR198 pKa = 11.84RR199 pKa = 11.84WHH201 pKa = 6.44HH202 pKa = 5.72VIKK205 pKa = 10.89DD206 pKa = 3.34GFEE209 pKa = 3.86RR210 pKa = 11.84HH211 pKa = 5.95AHH213 pKa = 6.02LKK215 pKa = 11.12DD216 pKa = 3.29NDD218 pKa = 4.02PYY220 pKa = 10.96FQNKK224 pKa = 9.72YY225 pKa = 8.89IFPMLLHH232 pKa = 7.04AKK234 pKa = 8.27TAIVKK239 pKa = 10.28KK240 pKa = 10.44DD241 pKa = 3.95DD242 pKa = 4.07PNKK245 pKa = 9.8MRR247 pKa = 11.84TIWGCSKK254 pKa = 10.23PWIIADD260 pKa = 3.27AMLYY264 pKa = 9.31WEE266 pKa = 4.52YY267 pKa = 10.78IAWIKK272 pKa = 10.3LNPGLTPMLWGYY284 pKa = 7.79EE285 pKa = 4.05TMTGGWMRR293 pKa = 11.84LNAALFSSYY302 pKa = 11.1LNKK305 pKa = 10.48CYY307 pKa = 9.66LTLDD311 pKa = 3.49WSRR314 pKa = 11.84FDD316 pKa = 3.04KK317 pKa = 10.71RR318 pKa = 11.84AYY320 pKa = 9.91FSIIRR325 pKa = 11.84RR326 pKa = 11.84IMFGILTYY334 pKa = 10.95LDD336 pKa = 3.84FDD338 pKa = 3.65NGYY341 pKa = 9.48VPNVNYY347 pKa = 9.95PDD349 pKa = 3.94TKK351 pKa = 11.01SDD353 pKa = 3.09WSHH356 pKa = 6.63LKK358 pKa = 9.46SQRR361 pKa = 11.84ILRR364 pKa = 11.84LWLWTLDD371 pKa = 3.69NLFNAPIVLPDD382 pKa = 2.78GRR384 pKa = 11.84MYY386 pKa = 11.0KK387 pKa = 10.2RR388 pKa = 11.84LFAGIPSGLFITQLLDD404 pKa = 2.7SWYY407 pKa = 9.6NYY409 pKa = 9.02TMIATILRR417 pKa = 11.84AMGFNPRR424 pKa = 11.84QCIIKK429 pKa = 9.98VQGDD433 pKa = 3.6DD434 pKa = 3.9SIVRR438 pKa = 11.84LYY440 pKa = 11.33VLIPPAKK447 pKa = 10.39HH448 pKa = 6.2EE449 pKa = 4.37DD450 pKa = 3.29FLLRR454 pKa = 11.84MQEE457 pKa = 4.0LADD460 pKa = 4.14HH461 pKa = 6.04YY462 pKa = 9.39FKK464 pKa = 11.25SVISVDD470 pKa = 3.1KK471 pKa = 11.48SEE473 pKa = 5.01IGSSLNGRR481 pKa = 11.84EE482 pKa = 3.88VLSYY486 pKa = 10.61RR487 pKa = 11.84NKK489 pKa = 10.43NGFPFRR495 pKa = 11.84DD496 pKa = 4.15EE497 pKa = 4.08IKK499 pKa = 10.24MIAQFYY505 pKa = 7.08HH506 pKa = 5.15TKK508 pKa = 10.25ARR510 pKa = 11.84KK511 pKa = 7.28STPEE515 pKa = 3.28IAMAQAIGFAYY526 pKa = 10.15ASCGNHH532 pKa = 6.52DD533 pKa = 3.93RR534 pKa = 11.84VLSVLEE540 pKa = 4.37DD541 pKa = 3.54MYY543 pKa = 11.84DD544 pKa = 3.2HH545 pKa = 6.78YY546 pKa = 11.29LSQGYY551 pKa = 9.3SPNPAGLTMVFGNSPDD567 pKa = 3.64TEE569 pKa = 4.2RR570 pKa = 11.84PFIDD574 pKa = 3.4MDD576 pKa = 3.85HH577 pKa = 6.77FPSISEE583 pKa = 3.6IKK585 pKa = 10.34RR586 pKa = 11.84FLVCTDD592 pKa = 3.1YY593 pKa = 11.58RR594 pKa = 11.84NDD596 pKa = 3.39VQSRR600 pKa = 11.84RR601 pKa = 11.84TWPPDD606 pKa = 3.43YY607 pKa = 10.28FLHH610 pKa = 7.08PPACKK615 pKa = 10.12PEE617 pKa = 3.73
Molecular weight: 72.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1102 |
485 |
617 |
551.0 |
63.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.534 ± 1.195 | 1.089 ± 0.164 |
6.171 ± 0.761 | 4.265 ± 0.473 |
5.717 ± 0.222 | 4.537 ± 0.384 |
3.267 ± 0.108 | 5.989 ± 0.519 |
4.174 ± 1.058 | 7.804 ± 0.366 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.632 ± 0.868 | 5.445 ± 0.333 |
7.441 ± 0.502 | 3.448 ± 0.549 |
5.808 ± 0.535 | 7.441 ± 1.015 |
6.443 ± 1.508 | 4.991 ± 0.743 |
1.996 ± 0.473 | 4.809 ± 0.683 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
