
Lentzea albidocapillata
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Lentzea
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
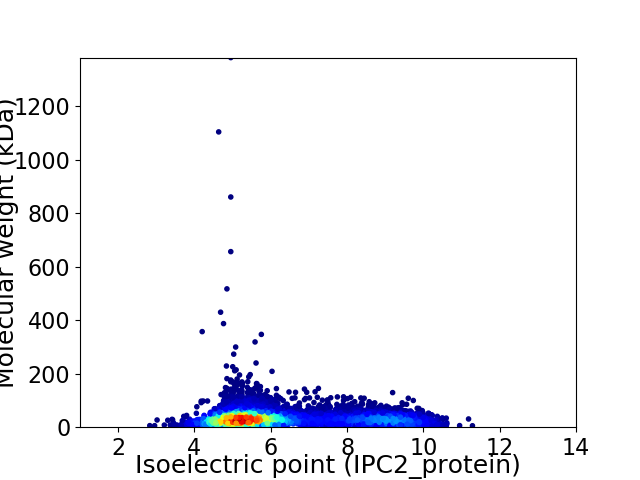
Virtual 2D-PAGE plot for 8155 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W2F6X4|A0A1W2F6X4_9PSEU Polar amino acid transport system substrate-binding protein OS=Lentzea albidocapillata OX=40571 GN=SAMN05660733_05210 PE=4 SV=1
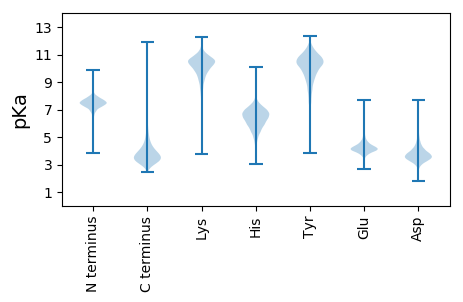
MM1 pKa = 7.77RR2 pKa = 11.84KK3 pKa = 9.01NVKK6 pKa = 9.24TGLLAAAVATGLVLAGPTASANTSAFHH33 pKa = 6.32PFGMGGTEE41 pKa = 4.42HH42 pKa = 6.01QMAWEE47 pKa = 4.21WFSHH51 pKa = 5.04QAAAYY56 pKa = 8.61AAAGHH61 pKa = 6.6GGHH64 pKa = 6.42SGDD67 pKa = 4.62GGDD70 pKa = 4.13AGSANTATTGDD81 pKa = 4.04SGDD84 pKa = 4.27SGDD87 pKa = 5.46SGDD90 pKa = 4.18TGHH93 pKa = 6.59NSAWVMCVHH102 pKa = 6.04SHH104 pKa = 6.89CEE106 pKa = 3.65ADD108 pKa = 3.84ADD110 pKa = 4.48SGDD113 pKa = 4.1SGDD116 pKa = 3.99TSTGDD121 pKa = 3.09TGNAANNSSTNGGDD135 pKa = 3.23TGHH138 pKa = 7.01GGNGGDD144 pKa = 4.31ADD146 pKa = 4.26ADD148 pKa = 3.85AEE150 pKa = 4.34NAVATIWDD158 pKa = 3.74EE159 pKa = 4.46DD160 pKa = 4.29GEE162 pKa = 5.22HH163 pKa = 6.83GDD165 pKa = 4.01EE166 pKa = 5.06DD167 pKa = 3.74NSSAHH172 pKa = 6.5SEE174 pKa = 4.15DD175 pKa = 3.79STHH178 pKa = 7.07GDD180 pKa = 3.55DD181 pKa = 4.18STHH184 pKa = 6.65HH185 pKa = 6.95GDD187 pKa = 3.69STHH190 pKa = 6.81HH191 pKa = 6.9GDD193 pKa = 3.89STHH196 pKa = 6.9HH197 pKa = 6.95GDD199 pKa = 4.36NEE201 pKa = 4.17ATASATAGHH210 pKa = 6.7GGDD213 pKa = 4.21SGDD216 pKa = 3.9GGDD219 pKa = 3.78AVSSNTATSGDD230 pKa = 3.89SGDD233 pKa = 4.47SGDD236 pKa = 5.46SGDD239 pKa = 4.18TGHH242 pKa = 6.59NSAWVMCVHH251 pKa = 6.84SDD253 pKa = 4.14CEE255 pKa = 4.26VGTDD259 pKa = 3.97SGDD262 pKa = 3.64SGDD265 pKa = 3.99TSTGDD270 pKa = 3.09TGNAANNSSTNGGDD284 pKa = 3.23TGHH287 pKa = 7.01GGNGGDD293 pKa = 3.9ADD295 pKa = 4.02AAAGTSTAVEE305 pKa = 3.88WDD307 pKa = 3.45DD308 pKa = 5.64DD309 pKa = 4.09EE310 pKa = 5.0NAAHH314 pKa = 7.07GDD316 pKa = 3.93SEE318 pKa = 4.33ATAAATAGNGGSSGAGGDD336 pKa = 3.28AVSVNTATTGDD347 pKa = 3.94SGDD350 pKa = 4.31SGDD353 pKa = 5.21SGNTGHH359 pKa = 6.66NSAWVMCAHH368 pKa = 7.13SICLTSTDD376 pKa = 4.32SGDD379 pKa = 4.03SGDD382 pKa = 3.99TSTGDD387 pKa = 3.12TGNATNNSSTNGGDD401 pKa = 3.23TGHH404 pKa = 7.01GGNGGDD410 pKa = 4.26ADD412 pKa = 5.52AMATDD417 pKa = 4.52LNEE420 pKa = 4.63GEE422 pKa = 5.09DD423 pKa = 3.89DD424 pKa = 3.63
MM1 pKa = 7.77RR2 pKa = 11.84KK3 pKa = 9.01NVKK6 pKa = 9.24TGLLAAAVATGLVLAGPTASANTSAFHH33 pKa = 6.32PFGMGGTEE41 pKa = 4.42HH42 pKa = 6.01QMAWEE47 pKa = 4.21WFSHH51 pKa = 5.04QAAAYY56 pKa = 8.61AAAGHH61 pKa = 6.6GGHH64 pKa = 6.42SGDD67 pKa = 4.62GGDD70 pKa = 4.13AGSANTATTGDD81 pKa = 4.04SGDD84 pKa = 4.27SGDD87 pKa = 5.46SGDD90 pKa = 4.18TGHH93 pKa = 6.59NSAWVMCVHH102 pKa = 6.04SHH104 pKa = 6.89CEE106 pKa = 3.65ADD108 pKa = 3.84ADD110 pKa = 4.48SGDD113 pKa = 4.1SGDD116 pKa = 3.99TSTGDD121 pKa = 3.09TGNAANNSSTNGGDD135 pKa = 3.23TGHH138 pKa = 7.01GGNGGDD144 pKa = 4.31ADD146 pKa = 4.26ADD148 pKa = 3.85AEE150 pKa = 4.34NAVATIWDD158 pKa = 3.74EE159 pKa = 4.46DD160 pKa = 4.29GEE162 pKa = 5.22HH163 pKa = 6.83GDD165 pKa = 4.01EE166 pKa = 5.06DD167 pKa = 3.74NSSAHH172 pKa = 6.5SEE174 pKa = 4.15DD175 pKa = 3.79STHH178 pKa = 7.07GDD180 pKa = 3.55DD181 pKa = 4.18STHH184 pKa = 6.65HH185 pKa = 6.95GDD187 pKa = 3.69STHH190 pKa = 6.81HH191 pKa = 6.9GDD193 pKa = 3.89STHH196 pKa = 6.9HH197 pKa = 6.95GDD199 pKa = 4.36NEE201 pKa = 4.17ATASATAGHH210 pKa = 6.7GGDD213 pKa = 4.21SGDD216 pKa = 3.9GGDD219 pKa = 3.78AVSSNTATSGDD230 pKa = 3.89SGDD233 pKa = 4.47SGDD236 pKa = 5.46SGDD239 pKa = 4.18TGHH242 pKa = 6.59NSAWVMCVHH251 pKa = 6.84SDD253 pKa = 4.14CEE255 pKa = 4.26VGTDD259 pKa = 3.97SGDD262 pKa = 3.64SGDD265 pKa = 3.99TSTGDD270 pKa = 3.09TGNAANNSSTNGGDD284 pKa = 3.23TGHH287 pKa = 7.01GGNGGDD293 pKa = 3.9ADD295 pKa = 4.02AAAGTSTAVEE305 pKa = 3.88WDD307 pKa = 3.45DD308 pKa = 5.64DD309 pKa = 4.09EE310 pKa = 5.0NAAHH314 pKa = 7.07GDD316 pKa = 3.93SEE318 pKa = 4.33ATAAATAGNGGSSGAGGDD336 pKa = 3.28AVSVNTATTGDD347 pKa = 3.94SGDD350 pKa = 4.31SGDD353 pKa = 5.21SGNTGHH359 pKa = 6.66NSAWVMCAHH368 pKa = 7.13SICLTSTDD376 pKa = 4.32SGDD379 pKa = 4.03SGDD382 pKa = 3.99TSTGDD387 pKa = 3.12TGNATNNSSTNGGDD401 pKa = 3.23TGHH404 pKa = 7.01GGNGGDD410 pKa = 4.26ADD412 pKa = 5.52AMATDD417 pKa = 4.52LNEE420 pKa = 4.63GEE422 pKa = 5.09DD423 pKa = 3.89DD424 pKa = 3.63
Molecular weight: 40.52 kDa
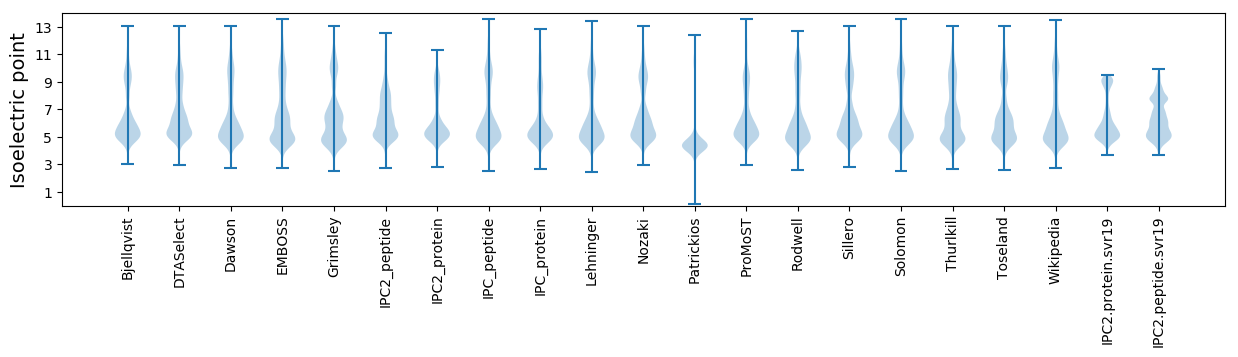
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W2EP90|A0A1W2EP90_9PSEU Alpha-galactosidase OS=Lentzea albidocapillata OX=40571 GN=SAMN05660733_04216 PE=3 SV=1
MM1 pKa = 7.53SKK3 pKa = 10.53GKK5 pKa = 8.66RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AKK17 pKa = 8.7THH19 pKa = 5.15GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AILAARR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.82GRR42 pKa = 11.84ATLSAA47 pKa = 4.08
MM1 pKa = 7.53SKK3 pKa = 10.53GKK5 pKa = 8.66RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AKK17 pKa = 8.7THH19 pKa = 5.15GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AILAARR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.82GRR42 pKa = 11.84ATLSAA47 pKa = 4.08
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2634138 |
39 |
12847 |
323.0 |
34.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.514 ± 0.035 | 0.797 ± 0.009 |
5.942 ± 0.025 | 5.614 ± 0.024 |
3.043 ± 0.016 | 8.871 ± 0.027 |
2.245 ± 0.014 | 3.605 ± 0.021 |
2.522 ± 0.026 | 10.416 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.847 ± 0.012 | 2.229 ± 0.025 |
5.549 ± 0.024 | 3.029 ± 0.019 |
7.457 ± 0.032 | 5.518 ± 0.019 |
6.047 ± 0.031 | 9.154 ± 0.033 |
1.589 ± 0.013 | 2.011 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
