
Porcine stool-associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; unclassified Porprismacovirus
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
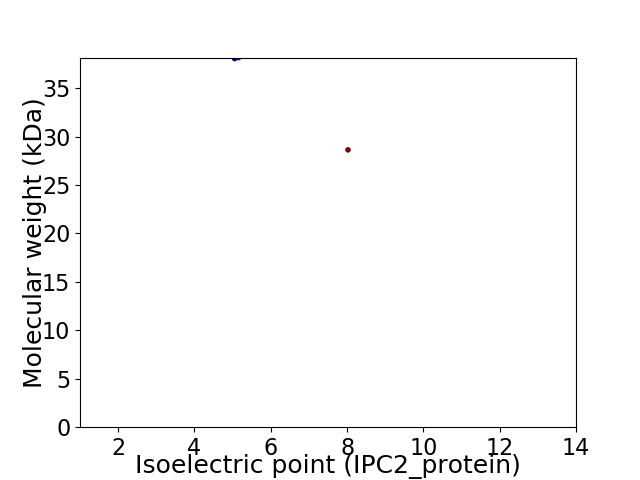
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
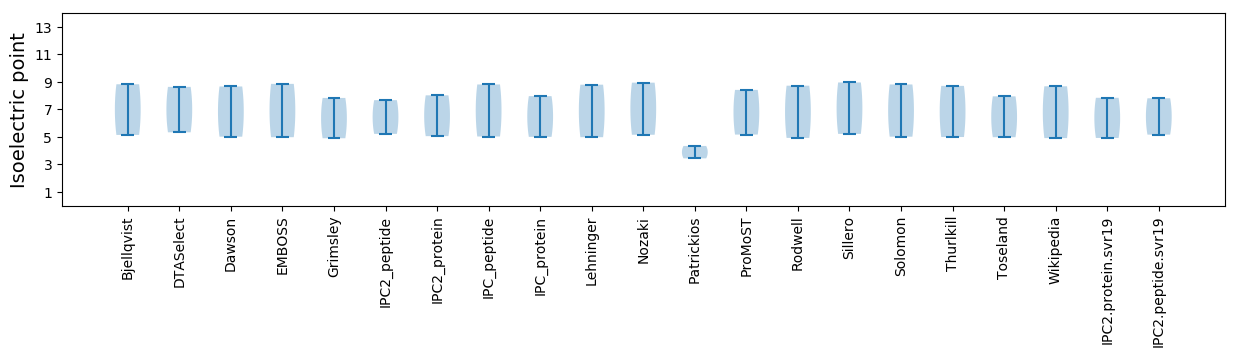
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A341ZMI4|A0A341ZMI4_9VIRU Putative capsid protein OS=Porcine stool-associated circular virus OX=1843773 PE=4 SV=1
MM1 pKa = 6.78ATNYY5 pKa = 9.85VKK7 pKa = 10.65ARR9 pKa = 11.84YY10 pKa = 8.8QEE12 pKa = 4.35IYY14 pKa = 11.04DD15 pKa = 4.09FGTQAGKK22 pKa = 6.91TTILGVHH29 pKa = 6.28SPTGNKK35 pKa = 7.74VQEE38 pKa = 4.04MLGGFFRR45 pKa = 11.84QFRR48 pKa = 11.84KK49 pKa = 10.1YY50 pKa = 9.85RR51 pKa = 11.84YY52 pKa = 8.73SGCTVTMVPAAQLPADD68 pKa = 4.06PLQVSFEE75 pKa = 4.22AGALTIDD82 pKa = 4.33PRR84 pKa = 11.84DD85 pKa = 3.65MLNPILFHH93 pKa = 6.25GAHH96 pKa = 6.16GEE98 pKa = 4.24SLNEE102 pKa = 3.91ALNLIYY108 pKa = 9.83NTSTAEE114 pKa = 4.15MVDD117 pKa = 3.45GASVKK122 pKa = 10.16EE123 pKa = 3.93IRR125 pKa = 11.84DD126 pKa = 3.51QFSISSWGSEE136 pKa = 3.56NAYY139 pKa = 10.65YY140 pKa = 10.51SALCDD145 pKa = 3.51PSFKK149 pKa = 10.82KK150 pKa = 10.77FGIQSGAKK158 pKa = 8.81VNLRR162 pKa = 11.84PMVHH166 pKa = 6.0PVVLNHH172 pKa = 6.9AMLPSDD178 pKa = 4.84SGVEE182 pKa = 3.71YY183 pKa = 11.03DD184 pKa = 4.06NASNHH189 pKa = 6.03YY190 pKa = 10.91DD191 pKa = 3.35MASDD195 pKa = 4.12GVGQILGDD203 pKa = 3.58NRR205 pKa = 11.84IGQGAIGNIAEE216 pKa = 4.23VSGAEE221 pKa = 4.01AVSVVPRR228 pKa = 11.84QMFTSGLRR236 pKa = 11.84PLGWLNTTQFAAPINVPNGSTNIDD260 pKa = 3.0WFLSYY265 pKa = 10.97SYY267 pKa = 10.28IPKK270 pKa = 10.1IMMGVFILPPSYY282 pKa = 9.11TQEE285 pKa = 3.69MYY287 pKa = 10.42FRR289 pKa = 11.84LIINHH294 pKa = 5.37YY295 pKa = 10.77FEE297 pKa = 5.98FKK299 pKa = 10.75DD300 pKa = 4.02FSTALTYY307 pKa = 11.03SPDD310 pKa = 3.54GARR313 pKa = 11.84LVYY316 pKa = 10.36TEE318 pKa = 5.24TIPDD322 pKa = 3.85PSGGASLSSASLDD335 pKa = 3.67VEE337 pKa = 4.18NGTIAKK343 pKa = 6.66TTDD346 pKa = 2.97GVFF349 pKa = 3.16
MM1 pKa = 6.78ATNYY5 pKa = 9.85VKK7 pKa = 10.65ARR9 pKa = 11.84YY10 pKa = 8.8QEE12 pKa = 4.35IYY14 pKa = 11.04DD15 pKa = 4.09FGTQAGKK22 pKa = 6.91TTILGVHH29 pKa = 6.28SPTGNKK35 pKa = 7.74VQEE38 pKa = 4.04MLGGFFRR45 pKa = 11.84QFRR48 pKa = 11.84KK49 pKa = 10.1YY50 pKa = 9.85RR51 pKa = 11.84YY52 pKa = 8.73SGCTVTMVPAAQLPADD68 pKa = 4.06PLQVSFEE75 pKa = 4.22AGALTIDD82 pKa = 4.33PRR84 pKa = 11.84DD85 pKa = 3.65MLNPILFHH93 pKa = 6.25GAHH96 pKa = 6.16GEE98 pKa = 4.24SLNEE102 pKa = 3.91ALNLIYY108 pKa = 9.83NTSTAEE114 pKa = 4.15MVDD117 pKa = 3.45GASVKK122 pKa = 10.16EE123 pKa = 3.93IRR125 pKa = 11.84DD126 pKa = 3.51QFSISSWGSEE136 pKa = 3.56NAYY139 pKa = 10.65YY140 pKa = 10.51SALCDD145 pKa = 3.51PSFKK149 pKa = 10.82KK150 pKa = 10.77FGIQSGAKK158 pKa = 8.81VNLRR162 pKa = 11.84PMVHH166 pKa = 6.0PVVLNHH172 pKa = 6.9AMLPSDD178 pKa = 4.84SGVEE182 pKa = 3.71YY183 pKa = 11.03DD184 pKa = 4.06NASNHH189 pKa = 6.03YY190 pKa = 10.91DD191 pKa = 3.35MASDD195 pKa = 4.12GVGQILGDD203 pKa = 3.58NRR205 pKa = 11.84IGQGAIGNIAEE216 pKa = 4.23VSGAEE221 pKa = 4.01AVSVVPRR228 pKa = 11.84QMFTSGLRR236 pKa = 11.84PLGWLNTTQFAAPINVPNGSTNIDD260 pKa = 3.0WFLSYY265 pKa = 10.97SYY267 pKa = 10.28IPKK270 pKa = 10.1IMMGVFILPPSYY282 pKa = 9.11TQEE285 pKa = 3.69MYY287 pKa = 10.42FRR289 pKa = 11.84LIINHH294 pKa = 5.37YY295 pKa = 10.77FEE297 pKa = 5.98FKK299 pKa = 10.75DD300 pKa = 4.02FSTALTYY307 pKa = 11.03SPDD310 pKa = 3.54GARR313 pKa = 11.84LVYY316 pKa = 10.36TEE318 pKa = 5.24TIPDD322 pKa = 3.85PSGGASLSSASLDD335 pKa = 3.67VEE337 pKa = 4.18NGTIAKK343 pKa = 6.66TTDD346 pKa = 2.97GVFF349 pKa = 3.16
Molecular weight: 38.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A341ZMI4|A0A341ZMI4_9VIRU Putative capsid protein OS=Porcine stool-associated circular virus OX=1843773 PE=4 SV=1
MM1 pKa = 6.24QTYY4 pKa = 9.74MMTIPRR10 pKa = 11.84TVSKK14 pKa = 10.35RR15 pKa = 11.84ALRR18 pKa = 11.84IMIEE22 pKa = 4.22KK23 pKa = 10.03NDD25 pKa = 3.84CKK27 pKa = 10.6KK28 pKa = 8.7WTIGKK33 pKa = 9.42EE34 pKa = 3.81KK35 pKa = 10.49GKK37 pKa = 10.28NGYY40 pKa = 7.38EE41 pKa = 3.45HH42 pKa = 5.57WQIRR46 pKa = 11.84IEE48 pKa = 4.1TSNDD52 pKa = 3.27DD53 pKa = 3.58FFEE56 pKa = 3.99WMQDD60 pKa = 3.68HH61 pKa = 7.09IPTAHH66 pKa = 6.76IEE68 pKa = 4.17KK69 pKa = 10.0SDD71 pKa = 3.77SGVDD75 pKa = 3.14EE76 pKa = 4.55CRR78 pKa = 11.84YY79 pKa = 6.09EE80 pKa = 4.29TKK82 pKa = 10.18EE83 pKa = 4.03GQYY86 pKa = 8.7VTYY89 pKa = 10.42SDD91 pKa = 4.24RR92 pKa = 11.84VQNLMQRR99 pKa = 11.84YY100 pKa = 8.21GAFRR104 pKa = 11.84PNQKK108 pKa = 9.94RR109 pKa = 11.84AMQALEE115 pKa = 4.08ATNDD119 pKa = 3.53RR120 pKa = 11.84QVVVWYY126 pKa = 10.21DD127 pKa = 3.2EE128 pKa = 4.14TGNVGKK134 pKa = 10.24SWFTGALWEE143 pKa = 4.56RR144 pKa = 11.84GLAYY148 pKa = 7.93VTPPTIDD155 pKa = 3.16TVKK158 pKa = 11.09GLIQWVASCYY168 pKa = 9.78IDD170 pKa = 4.44GGWRR174 pKa = 11.84PYY176 pKa = 11.0VIIDD180 pKa = 4.19VPRR183 pKa = 11.84SWKK186 pKa = 9.71WSEE189 pKa = 3.8QLYY192 pKa = 10.74SAIEE196 pKa = 4.22SIKK199 pKa = 11.0DD200 pKa = 3.09GLIYY204 pKa = 9.1DD205 pKa = 4.01TRR207 pKa = 11.84YY208 pKa = 8.56HH209 pKa = 6.86SRR211 pKa = 11.84MINIRR216 pKa = 11.84GVKK219 pKa = 9.03VLVMTNTMPKK229 pKa = 9.86LDD231 pKa = 4.6KK232 pKa = 10.62LSKK235 pKa = 10.6DD236 pKa = 3.0RR237 pKa = 11.84WCICTFF243 pKa = 3.64
MM1 pKa = 6.24QTYY4 pKa = 9.74MMTIPRR10 pKa = 11.84TVSKK14 pKa = 10.35RR15 pKa = 11.84ALRR18 pKa = 11.84IMIEE22 pKa = 4.22KK23 pKa = 10.03NDD25 pKa = 3.84CKK27 pKa = 10.6KK28 pKa = 8.7WTIGKK33 pKa = 9.42EE34 pKa = 3.81KK35 pKa = 10.49GKK37 pKa = 10.28NGYY40 pKa = 7.38EE41 pKa = 3.45HH42 pKa = 5.57WQIRR46 pKa = 11.84IEE48 pKa = 4.1TSNDD52 pKa = 3.27DD53 pKa = 3.58FFEE56 pKa = 3.99WMQDD60 pKa = 3.68HH61 pKa = 7.09IPTAHH66 pKa = 6.76IEE68 pKa = 4.17KK69 pKa = 10.0SDD71 pKa = 3.77SGVDD75 pKa = 3.14EE76 pKa = 4.55CRR78 pKa = 11.84YY79 pKa = 6.09EE80 pKa = 4.29TKK82 pKa = 10.18EE83 pKa = 4.03GQYY86 pKa = 8.7VTYY89 pKa = 10.42SDD91 pKa = 4.24RR92 pKa = 11.84VQNLMQRR99 pKa = 11.84YY100 pKa = 8.21GAFRR104 pKa = 11.84PNQKK108 pKa = 9.94RR109 pKa = 11.84AMQALEE115 pKa = 4.08ATNDD119 pKa = 3.53RR120 pKa = 11.84QVVVWYY126 pKa = 10.21DD127 pKa = 3.2EE128 pKa = 4.14TGNVGKK134 pKa = 10.24SWFTGALWEE143 pKa = 4.56RR144 pKa = 11.84GLAYY148 pKa = 7.93VTPPTIDD155 pKa = 3.16TVKK158 pKa = 11.09GLIQWVASCYY168 pKa = 9.78IDD170 pKa = 4.44GGWRR174 pKa = 11.84PYY176 pKa = 11.0VIIDD180 pKa = 4.19VPRR183 pKa = 11.84SWKK186 pKa = 9.71WSEE189 pKa = 3.8QLYY192 pKa = 10.74SAIEE196 pKa = 4.22SIKK199 pKa = 11.0DD200 pKa = 3.09GLIYY204 pKa = 9.1DD205 pKa = 4.01TRR207 pKa = 11.84YY208 pKa = 8.56HH209 pKa = 6.86SRR211 pKa = 11.84MINIRR216 pKa = 11.84GVKK219 pKa = 9.03VLVMTNTMPKK229 pKa = 9.86LDD231 pKa = 4.6KK232 pKa = 10.62LSKK235 pKa = 10.6DD236 pKa = 3.0RR237 pKa = 11.84WCICTFF243 pKa = 3.64
Molecular weight: 28.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
592 |
243 |
349 |
296.0 |
33.34 |
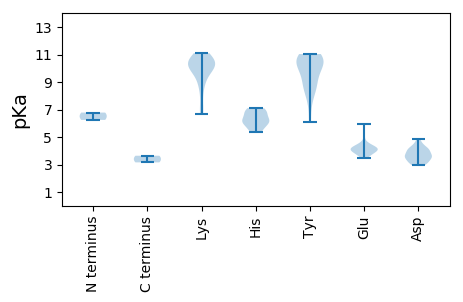
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.588 ± 1.58 | 1.182 ± 0.559 |
5.743 ± 0.538 | 4.899 ± 0.551 |
3.885 ± 1.168 | 7.77 ± 1.021 |
1.858 ± 0.136 | 6.757 ± 0.679 |
4.899 ± 1.603 | 6.081 ± 0.993 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.716 ± 0.255 | 4.73 ± 0.656 |
4.73 ± 0.919 | 4.054 ± 0.302 |
4.899 ± 1.34 | 7.432 ± 1.331 |
6.757 ± 0.416 | 6.588 ± 0.002 |
2.365 ± 1.382 | 5.068 ± 0.18 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
