
Haplochromis burtoni (Burton s mouthbrooder) (Chromis burtoni)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia;
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
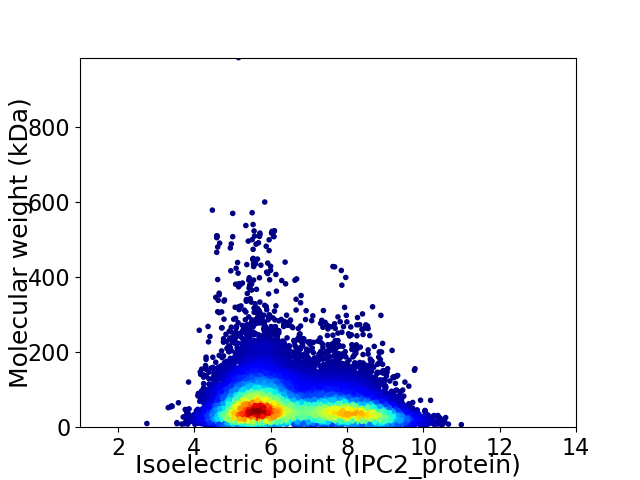
Virtual 2D-PAGE plot for 34332 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3Q2W2Y3|A0A3Q2W2Y3_HAPBU Tripartite motif-containing protein 16-like OS=Haplochromis burtoni OX=8153 PE=4 SV=1
MM1 pKa = 7.23PVIWEE6 pKa = 4.22LVSTDD11 pKa = 2.95TNAWSKK17 pKa = 11.54SFGQSLINTFVMYY30 pKa = 10.76LEE32 pKa = 4.8NSEE35 pKa = 4.25QNKK38 pKa = 7.16GTRR41 pKa = 11.84EE42 pKa = 4.23GEE44 pKa = 4.06PSTEE48 pKa = 3.85AMFAEE53 pKa = 4.84QCQVPRR59 pKa = 11.84LTEE62 pKa = 4.04AADD65 pKa = 3.49LTGPSKK71 pKa = 11.0GSAEE75 pKa = 4.24LGVKK79 pKa = 9.95VLLQEE84 pKa = 4.1YY85 pKa = 9.02TEE87 pKa = 4.13QAVSKK92 pKa = 10.25EE93 pKa = 4.13MDD95 pKa = 3.55PSIPCLEE102 pKa = 4.03QCNLNSDD109 pKa = 4.0EE110 pKa = 5.64LEE112 pKa = 3.99SCTYY116 pKa = 10.61FNGHH120 pKa = 6.83ADD122 pKa = 3.14SFEE125 pKa = 4.21QYY127 pKa = 10.43SEE129 pKa = 4.38SNGFFEE135 pKa = 6.0SDD137 pKa = 3.58QILDD141 pKa = 3.47KK142 pKa = 11.57CEE144 pKa = 3.91TSEE147 pKa = 5.08LSQPCDD153 pKa = 3.0EE154 pKa = 4.71CTQHH158 pKa = 6.77CEE160 pKa = 4.02CFKK163 pKa = 11.06SSEE166 pKa = 4.06HH167 pKa = 6.62CGNHH171 pKa = 5.74SSASEE176 pKa = 3.93CSACCEE182 pKa = 3.78QCAEE186 pKa = 4.28HH187 pKa = 6.65LQLFQKK193 pKa = 10.22CKK195 pKa = 10.34PSDD198 pKa = 3.58QQLEE202 pKa = 4.16ISDD205 pKa = 4.46LDD207 pKa = 3.88ADD209 pKa = 4.11EE210 pKa = 6.22LEE212 pKa = 4.71VNCGSLGLCEE222 pKa = 4.7RR223 pKa = 11.84CDD225 pKa = 4.18FNLEE229 pKa = 4.26CTDD232 pKa = 3.76HH233 pKa = 7.43PEE235 pKa = 4.26ALQQYY240 pKa = 8.3EE241 pKa = 4.3PSQQQSDD248 pKa = 4.37CFDD251 pKa = 4.08SEE253 pKa = 5.04SDD255 pKa = 3.16TSTEE259 pKa = 3.88NFQQCEE265 pKa = 4.25TVDD268 pKa = 3.79SLNCNPEE275 pKa = 3.53VCEE278 pKa = 3.77YY279 pKa = 11.4DD280 pKa = 3.88EE281 pKa = 5.92ISDD284 pKa = 3.62EE285 pKa = 4.6TEE287 pKa = 3.79FTNDD291 pKa = 3.27EE292 pKa = 4.21DD293 pKa = 5.13DD294 pKa = 4.36EE295 pKa = 4.34EE296 pKa = 6.33AEE298 pKa = 4.16EE299 pKa = 3.92EE300 pKa = 5.0DD301 pKa = 5.82YY302 pKa = 11.87YY303 pKa = 11.62DD304 pKa = 6.04DD305 pKa = 6.37DD306 pKa = 7.33DD307 pKa = 7.3GDD309 pKa = 4.4FEE311 pKa = 7.75DD312 pKa = 5.23DD313 pKa = 3.66LSYY316 pKa = 10.92KK317 pKa = 10.48AKK319 pKa = 10.45QNEE322 pKa = 4.51TEE324 pKa = 4.23FSDD327 pKa = 4.21EE328 pKa = 4.16EE329 pKa = 4.45SHH331 pKa = 7.3SSSEE335 pKa = 4.07DD336 pKa = 3.31TPVQVFFDD344 pKa = 3.72DD345 pKa = 4.07TEE347 pKa = 4.28NVAFATDD354 pKa = 2.88LWEE357 pKa = 3.89THH359 pKa = 6.43EE360 pKa = 4.12YY361 pKa = 9.38TKK363 pKa = 11.21EE364 pKa = 3.91DD365 pKa = 3.59FQQALTTEE373 pKa = 4.51VPTEE377 pKa = 4.02SGEE380 pKa = 4.07PCGNDD385 pKa = 3.19DD386 pKa = 4.36YY387 pKa = 11.68EE388 pKa = 4.77VSQEE392 pKa = 3.88PAQQYY397 pKa = 7.64STSEE401 pKa = 4.1QYY403 pKa = 10.73KK404 pKa = 9.64SEE406 pKa = 4.18SSGEE410 pKa = 3.88EE411 pKa = 4.31DD412 pKa = 4.12GSSDD416 pKa = 3.65CSSVEE421 pKa = 3.91TKK423 pKa = 10.55SFKK426 pKa = 9.88TCPEE430 pKa = 3.65GSIPSDD436 pKa = 3.43PFSDD440 pKa = 4.01SSEE443 pKa = 4.21EE444 pKa = 4.08SDD446 pKa = 4.85KK447 pKa = 11.47GPQEE451 pKa = 4.46DD452 pKa = 4.29SSDD455 pKa = 3.61EE456 pKa = 3.97QTEE459 pKa = 4.3WEE461 pKa = 4.27SFEE464 pKa = 5.06DD465 pKa = 4.57EE466 pKa = 4.46EE467 pKa = 4.97VQQSDD472 pKa = 3.67EE473 pKa = 4.38NKK475 pKa = 10.4SKK477 pKa = 9.59TVDD480 pKa = 3.44VTVEE484 pKa = 4.23DD485 pKa = 4.37YY486 pKa = 11.32FDD488 pKa = 5.6LFDD491 pKa = 3.83KK492 pKa = 10.82VDD494 pKa = 3.73YY495 pKa = 9.85YY496 pKa = 11.7GHH498 pKa = 7.52IFTQKK503 pKa = 9.46QRR505 pKa = 11.84YY506 pKa = 8.15ISCFDD511 pKa = 3.46GGDD514 pKa = 3.48IHH516 pKa = 7.4DD517 pKa = 4.23HH518 pKa = 6.7LYY520 pKa = 11.08LEE522 pKa = 4.83EE523 pKa = 3.89EE524 pKa = 4.31AKK526 pKa = 10.18KK527 pKa = 10.2HH528 pKa = 5.23KK529 pKa = 10.76AKK531 pKa = 9.68TLYY534 pKa = 10.18QFEE537 pKa = 5.1DD538 pKa = 3.19INRR541 pKa = 11.84EE542 pKa = 4.08ANVKK546 pKa = 8.94DD547 pKa = 3.51TDD549 pKa = 3.85TCSEE553 pKa = 4.31DD554 pKa = 3.63ASEE557 pKa = 4.08DD558 pKa = 3.68TYY560 pKa = 11.9EE561 pKa = 4.4EE562 pKa = 4.28VDD564 pKa = 3.24VDD566 pKa = 4.89QSDD569 pKa = 4.35GSCDD573 pKa = 4.76LEE575 pKa = 4.5TQSEE579 pKa = 4.23DD580 pKa = 2.83WLIAKK585 pKa = 9.83SEE587 pKa = 4.34SSFEE591 pKa = 4.05EE592 pKa = 4.29DD593 pKa = 5.07DD594 pKa = 3.91EE595 pKa = 5.45SKK597 pKa = 10.06TEE599 pKa = 3.91NGVVYY604 pKa = 10.82AEE606 pKa = 4.07NDD608 pKa = 3.08EE609 pKa = 4.22TAEE612 pKa = 4.29DD613 pKa = 3.77DD614 pKa = 3.84KK615 pKa = 12.04APIDD619 pKa = 4.01EE620 pKa = 4.58EE621 pKa = 4.95ACASDD626 pKa = 3.53DD627 pKa = 3.95HH628 pKa = 8.44VSEE631 pKa = 4.28EE632 pKa = 4.52AEE634 pKa = 4.25VRR636 pKa = 11.84PSTGPEE642 pKa = 3.5GSMFAPLAKK651 pKa = 9.86EE652 pKa = 3.72ISVEE656 pKa = 3.59GDD658 pKa = 2.9AYY660 pKa = 10.61EE661 pKa = 6.01DD662 pKa = 3.72ILCSSHH668 pKa = 6.71YY669 pKa = 10.5CEE671 pKa = 5.27PKK673 pKa = 10.4PSATDD678 pKa = 3.32EE679 pKa = 4.27TASITDD685 pKa = 3.39YY686 pKa = 11.59TEE688 pKa = 4.17DD689 pKa = 4.19DD690 pKa = 3.62KK691 pKa = 11.98AKK693 pKa = 10.36PEE695 pKa = 4.34PEE697 pKa = 4.2DD698 pKa = 4.52DD699 pKa = 4.17VFITCSEE706 pKa = 4.16KK707 pKa = 10.35EE708 pKa = 4.48PYY710 pKa = 9.08WSLIDD715 pKa = 3.76NDD717 pKa = 4.22EE718 pKa = 3.85NRR720 pKa = 11.84QLCEE724 pKa = 4.1PGVEE728 pKa = 4.08EE729 pKa = 5.11YY730 pKa = 10.9YY731 pKa = 10.88VYY733 pKa = 10.38QIKK736 pKa = 10.5SIQSSFKK743 pKa = 9.49QALNGFILEE752 pKa = 4.4RR753 pKa = 11.84NSHH756 pKa = 4.69NQITNDD762 pKa = 3.35EE763 pKa = 4.19NASRR767 pKa = 11.84SQSQDD772 pKa = 2.59ALISLKK778 pKa = 10.4EE779 pKa = 3.75LAATGLCPEE788 pKa = 4.66EE789 pKa = 4.57CVSDD793 pKa = 3.43TFGINDD799 pKa = 4.1VIDD802 pKa = 3.93MNSSAVEE809 pKa = 4.09EE810 pKa = 4.22EE811 pKa = 4.49CVLNEE816 pKa = 3.97ISRR819 pKa = 11.84EE820 pKa = 3.95TGPPVGIIHH829 pKa = 6.49SVQEE833 pKa = 3.84RR834 pKa = 11.84DD835 pKa = 3.35MVLMKK840 pKa = 10.74TEE842 pKa = 4.82DD843 pKa = 3.77NNDD846 pKa = 3.48SEE848 pKa = 4.58QSRR851 pKa = 11.84EE852 pKa = 3.97SDD854 pKa = 3.12EE855 pKa = 4.82DD856 pKa = 3.63QSDD859 pKa = 3.83DD860 pKa = 3.76EE861 pKa = 5.11SYY863 pKa = 8.0EE864 pKa = 4.06TCEE867 pKa = 4.22CDD869 pKa = 3.49YY870 pKa = 11.01CITPTEE876 pKa = 4.13EE877 pKa = 4.62LPGEE881 pKa = 4.06PLLPQMEE888 pKa = 4.81SNDD891 pKa = 3.5EE892 pKa = 3.98GKK894 pKa = 9.71ICVVIDD900 pKa = 3.71LDD902 pKa = 3.74EE903 pKa = 4.47TLVHH907 pKa = 7.06SSFKK911 pKa = 10.36PVNNADD917 pKa = 4.05FIIPVEE923 pKa = 3.92IDD925 pKa = 3.4GTVHH929 pKa = 5.33QVYY932 pKa = 9.86VLKK935 pKa = 10.52RR936 pKa = 11.84PHH938 pKa = 7.15VDD940 pKa = 2.95EE941 pKa = 4.17FLKK944 pKa = 10.97RR945 pKa = 11.84MGEE948 pKa = 3.93MFEE951 pKa = 4.9CVLFTASLSKK961 pKa = 11.06YY962 pKa = 9.64ADD964 pKa = 3.7PVSDD968 pKa = 6.39LLDD971 pKa = 2.94KK972 pKa = 10.75WGAFRR977 pKa = 11.84SRR979 pKa = 11.84LFRR982 pKa = 11.84EE983 pKa = 3.88ACVFHH988 pKa = 6.91KK989 pKa = 10.84GNYY992 pKa = 9.11VKK994 pKa = 10.65DD995 pKa = 3.95LSRR998 pKa = 11.84LGRR1001 pKa = 11.84DD1002 pKa = 3.3LNKK1005 pKa = 10.22VIILDD1010 pKa = 3.92NSPASYY1016 pKa = 10.24IFHH1019 pKa = 7.73PEE1021 pKa = 3.29NAVPVASWFNDD1032 pKa = 3.14MSDD1035 pKa = 3.64TEE1037 pKa = 4.74LLDD1040 pKa = 4.78LIPFFEE1046 pKa = 4.75RR1047 pKa = 11.84LSKK1050 pKa = 10.91VDD1052 pKa = 4.9DD1053 pKa = 4.23IYY1055 pKa = 11.42DD1056 pKa = 3.65ILKK1059 pKa = 9.52HH1060 pKa = 5.91QSTSQSS1066 pKa = 3.06
MM1 pKa = 7.23PVIWEE6 pKa = 4.22LVSTDD11 pKa = 2.95TNAWSKK17 pKa = 11.54SFGQSLINTFVMYY30 pKa = 10.76LEE32 pKa = 4.8NSEE35 pKa = 4.25QNKK38 pKa = 7.16GTRR41 pKa = 11.84EE42 pKa = 4.23GEE44 pKa = 4.06PSTEE48 pKa = 3.85AMFAEE53 pKa = 4.84QCQVPRR59 pKa = 11.84LTEE62 pKa = 4.04AADD65 pKa = 3.49LTGPSKK71 pKa = 11.0GSAEE75 pKa = 4.24LGVKK79 pKa = 9.95VLLQEE84 pKa = 4.1YY85 pKa = 9.02TEE87 pKa = 4.13QAVSKK92 pKa = 10.25EE93 pKa = 4.13MDD95 pKa = 3.55PSIPCLEE102 pKa = 4.03QCNLNSDD109 pKa = 4.0EE110 pKa = 5.64LEE112 pKa = 3.99SCTYY116 pKa = 10.61FNGHH120 pKa = 6.83ADD122 pKa = 3.14SFEE125 pKa = 4.21QYY127 pKa = 10.43SEE129 pKa = 4.38SNGFFEE135 pKa = 6.0SDD137 pKa = 3.58QILDD141 pKa = 3.47KK142 pKa = 11.57CEE144 pKa = 3.91TSEE147 pKa = 5.08LSQPCDD153 pKa = 3.0EE154 pKa = 4.71CTQHH158 pKa = 6.77CEE160 pKa = 4.02CFKK163 pKa = 11.06SSEE166 pKa = 4.06HH167 pKa = 6.62CGNHH171 pKa = 5.74SSASEE176 pKa = 3.93CSACCEE182 pKa = 3.78QCAEE186 pKa = 4.28HH187 pKa = 6.65LQLFQKK193 pKa = 10.22CKK195 pKa = 10.34PSDD198 pKa = 3.58QQLEE202 pKa = 4.16ISDD205 pKa = 4.46LDD207 pKa = 3.88ADD209 pKa = 4.11EE210 pKa = 6.22LEE212 pKa = 4.71VNCGSLGLCEE222 pKa = 4.7RR223 pKa = 11.84CDD225 pKa = 4.18FNLEE229 pKa = 4.26CTDD232 pKa = 3.76HH233 pKa = 7.43PEE235 pKa = 4.26ALQQYY240 pKa = 8.3EE241 pKa = 4.3PSQQQSDD248 pKa = 4.37CFDD251 pKa = 4.08SEE253 pKa = 5.04SDD255 pKa = 3.16TSTEE259 pKa = 3.88NFQQCEE265 pKa = 4.25TVDD268 pKa = 3.79SLNCNPEE275 pKa = 3.53VCEE278 pKa = 3.77YY279 pKa = 11.4DD280 pKa = 3.88EE281 pKa = 5.92ISDD284 pKa = 3.62EE285 pKa = 4.6TEE287 pKa = 3.79FTNDD291 pKa = 3.27EE292 pKa = 4.21DD293 pKa = 5.13DD294 pKa = 4.36EE295 pKa = 4.34EE296 pKa = 6.33AEE298 pKa = 4.16EE299 pKa = 3.92EE300 pKa = 5.0DD301 pKa = 5.82YY302 pKa = 11.87YY303 pKa = 11.62DD304 pKa = 6.04DD305 pKa = 6.37DD306 pKa = 7.33DD307 pKa = 7.3GDD309 pKa = 4.4FEE311 pKa = 7.75DD312 pKa = 5.23DD313 pKa = 3.66LSYY316 pKa = 10.92KK317 pKa = 10.48AKK319 pKa = 10.45QNEE322 pKa = 4.51TEE324 pKa = 4.23FSDD327 pKa = 4.21EE328 pKa = 4.16EE329 pKa = 4.45SHH331 pKa = 7.3SSSEE335 pKa = 4.07DD336 pKa = 3.31TPVQVFFDD344 pKa = 3.72DD345 pKa = 4.07TEE347 pKa = 4.28NVAFATDD354 pKa = 2.88LWEE357 pKa = 3.89THH359 pKa = 6.43EE360 pKa = 4.12YY361 pKa = 9.38TKK363 pKa = 11.21EE364 pKa = 3.91DD365 pKa = 3.59FQQALTTEE373 pKa = 4.51VPTEE377 pKa = 4.02SGEE380 pKa = 4.07PCGNDD385 pKa = 3.19DD386 pKa = 4.36YY387 pKa = 11.68EE388 pKa = 4.77VSQEE392 pKa = 3.88PAQQYY397 pKa = 7.64STSEE401 pKa = 4.1QYY403 pKa = 10.73KK404 pKa = 9.64SEE406 pKa = 4.18SSGEE410 pKa = 3.88EE411 pKa = 4.31DD412 pKa = 4.12GSSDD416 pKa = 3.65CSSVEE421 pKa = 3.91TKK423 pKa = 10.55SFKK426 pKa = 9.88TCPEE430 pKa = 3.65GSIPSDD436 pKa = 3.43PFSDD440 pKa = 4.01SSEE443 pKa = 4.21EE444 pKa = 4.08SDD446 pKa = 4.85KK447 pKa = 11.47GPQEE451 pKa = 4.46DD452 pKa = 4.29SSDD455 pKa = 3.61EE456 pKa = 3.97QTEE459 pKa = 4.3WEE461 pKa = 4.27SFEE464 pKa = 5.06DD465 pKa = 4.57EE466 pKa = 4.46EE467 pKa = 4.97VQQSDD472 pKa = 3.67EE473 pKa = 4.38NKK475 pKa = 10.4SKK477 pKa = 9.59TVDD480 pKa = 3.44VTVEE484 pKa = 4.23DD485 pKa = 4.37YY486 pKa = 11.32FDD488 pKa = 5.6LFDD491 pKa = 3.83KK492 pKa = 10.82VDD494 pKa = 3.73YY495 pKa = 9.85YY496 pKa = 11.7GHH498 pKa = 7.52IFTQKK503 pKa = 9.46QRR505 pKa = 11.84YY506 pKa = 8.15ISCFDD511 pKa = 3.46GGDD514 pKa = 3.48IHH516 pKa = 7.4DD517 pKa = 4.23HH518 pKa = 6.7LYY520 pKa = 11.08LEE522 pKa = 4.83EE523 pKa = 3.89EE524 pKa = 4.31AKK526 pKa = 10.18KK527 pKa = 10.2HH528 pKa = 5.23KK529 pKa = 10.76AKK531 pKa = 9.68TLYY534 pKa = 10.18QFEE537 pKa = 5.1DD538 pKa = 3.19INRR541 pKa = 11.84EE542 pKa = 4.08ANVKK546 pKa = 8.94DD547 pKa = 3.51TDD549 pKa = 3.85TCSEE553 pKa = 4.31DD554 pKa = 3.63ASEE557 pKa = 4.08DD558 pKa = 3.68TYY560 pKa = 11.9EE561 pKa = 4.4EE562 pKa = 4.28VDD564 pKa = 3.24VDD566 pKa = 4.89QSDD569 pKa = 4.35GSCDD573 pKa = 4.76LEE575 pKa = 4.5TQSEE579 pKa = 4.23DD580 pKa = 2.83WLIAKK585 pKa = 9.83SEE587 pKa = 4.34SSFEE591 pKa = 4.05EE592 pKa = 4.29DD593 pKa = 5.07DD594 pKa = 3.91EE595 pKa = 5.45SKK597 pKa = 10.06TEE599 pKa = 3.91NGVVYY604 pKa = 10.82AEE606 pKa = 4.07NDD608 pKa = 3.08EE609 pKa = 4.22TAEE612 pKa = 4.29DD613 pKa = 3.77DD614 pKa = 3.84KK615 pKa = 12.04APIDD619 pKa = 4.01EE620 pKa = 4.58EE621 pKa = 4.95ACASDD626 pKa = 3.53DD627 pKa = 3.95HH628 pKa = 8.44VSEE631 pKa = 4.28EE632 pKa = 4.52AEE634 pKa = 4.25VRR636 pKa = 11.84PSTGPEE642 pKa = 3.5GSMFAPLAKK651 pKa = 9.86EE652 pKa = 3.72ISVEE656 pKa = 3.59GDD658 pKa = 2.9AYY660 pKa = 10.61EE661 pKa = 6.01DD662 pKa = 3.72ILCSSHH668 pKa = 6.71YY669 pKa = 10.5CEE671 pKa = 5.27PKK673 pKa = 10.4PSATDD678 pKa = 3.32EE679 pKa = 4.27TASITDD685 pKa = 3.39YY686 pKa = 11.59TEE688 pKa = 4.17DD689 pKa = 4.19DD690 pKa = 3.62KK691 pKa = 11.98AKK693 pKa = 10.36PEE695 pKa = 4.34PEE697 pKa = 4.2DD698 pKa = 4.52DD699 pKa = 4.17VFITCSEE706 pKa = 4.16KK707 pKa = 10.35EE708 pKa = 4.48PYY710 pKa = 9.08WSLIDD715 pKa = 3.76NDD717 pKa = 4.22EE718 pKa = 3.85NRR720 pKa = 11.84QLCEE724 pKa = 4.1PGVEE728 pKa = 4.08EE729 pKa = 5.11YY730 pKa = 10.9YY731 pKa = 10.88VYY733 pKa = 10.38QIKK736 pKa = 10.5SIQSSFKK743 pKa = 9.49QALNGFILEE752 pKa = 4.4RR753 pKa = 11.84NSHH756 pKa = 4.69NQITNDD762 pKa = 3.35EE763 pKa = 4.19NASRR767 pKa = 11.84SQSQDD772 pKa = 2.59ALISLKK778 pKa = 10.4EE779 pKa = 3.75LAATGLCPEE788 pKa = 4.66EE789 pKa = 4.57CVSDD793 pKa = 3.43TFGINDD799 pKa = 4.1VIDD802 pKa = 3.93MNSSAVEE809 pKa = 4.09EE810 pKa = 4.22EE811 pKa = 4.49CVLNEE816 pKa = 3.97ISRR819 pKa = 11.84EE820 pKa = 3.95TGPPVGIIHH829 pKa = 6.49SVQEE833 pKa = 3.84RR834 pKa = 11.84DD835 pKa = 3.35MVLMKK840 pKa = 10.74TEE842 pKa = 4.82DD843 pKa = 3.77NNDD846 pKa = 3.48SEE848 pKa = 4.58QSRR851 pKa = 11.84EE852 pKa = 3.97SDD854 pKa = 3.12EE855 pKa = 4.82DD856 pKa = 3.63QSDD859 pKa = 3.83DD860 pKa = 3.76EE861 pKa = 5.11SYY863 pKa = 8.0EE864 pKa = 4.06TCEE867 pKa = 4.22CDD869 pKa = 3.49YY870 pKa = 11.01CITPTEE876 pKa = 4.13EE877 pKa = 4.62LPGEE881 pKa = 4.06PLLPQMEE888 pKa = 4.81SNDD891 pKa = 3.5EE892 pKa = 3.98GKK894 pKa = 9.71ICVVIDD900 pKa = 3.71LDD902 pKa = 3.74EE903 pKa = 4.47TLVHH907 pKa = 7.06SSFKK911 pKa = 10.36PVNNADD917 pKa = 4.05FIIPVEE923 pKa = 3.92IDD925 pKa = 3.4GTVHH929 pKa = 5.33QVYY932 pKa = 9.86VLKK935 pKa = 10.52RR936 pKa = 11.84PHH938 pKa = 7.15VDD940 pKa = 2.95EE941 pKa = 4.17FLKK944 pKa = 10.97RR945 pKa = 11.84MGEE948 pKa = 3.93MFEE951 pKa = 4.9CVLFTASLSKK961 pKa = 11.06YY962 pKa = 9.64ADD964 pKa = 3.7PVSDD968 pKa = 6.39LLDD971 pKa = 2.94KK972 pKa = 10.75WGAFRR977 pKa = 11.84SRR979 pKa = 11.84LFRR982 pKa = 11.84EE983 pKa = 3.88ACVFHH988 pKa = 6.91KK989 pKa = 10.84GNYY992 pKa = 9.11VKK994 pKa = 10.65DD995 pKa = 3.95LSRR998 pKa = 11.84LGRR1001 pKa = 11.84DD1002 pKa = 3.3LNKK1005 pKa = 10.22VIILDD1010 pKa = 3.92NSPASYY1016 pKa = 10.24IFHH1019 pKa = 7.73PEE1021 pKa = 3.29NAVPVASWFNDD1032 pKa = 3.14MSDD1035 pKa = 3.64TEE1037 pKa = 4.74LLDD1040 pKa = 4.78LIPFFEE1046 pKa = 4.75RR1047 pKa = 11.84LSKK1050 pKa = 10.91VDD1052 pKa = 4.9DD1053 pKa = 4.23IYY1055 pKa = 11.42DD1056 pKa = 3.65ILKK1059 pKa = 9.52HH1060 pKa = 5.91QSTSQSS1066 pKa = 3.06
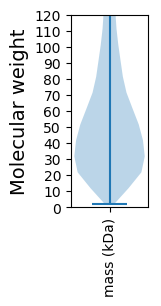
Molecular weight: 120.3 kDa
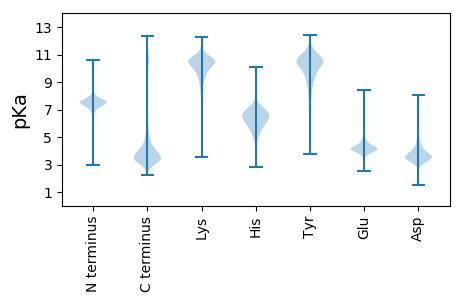
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q2WAV9|A0A3Q2WAV9_HAPBU PHD finger protein 23 OS=Haplochromis burtoni OX=8153 PE=4 SV=1
WW1 pKa = 7.07GLSPEE6 pKa = 4.09KK7 pKa = 10.27SSRR10 pKa = 11.84PFARR14 pKa = 11.84LRR16 pKa = 11.84EE17 pKa = 4.06PNRR20 pKa = 11.84IQEE23 pKa = 4.26AALFVPQPVRR33 pKa = 11.84SQILQWAHH41 pKa = 5.04TARR44 pKa = 11.84FSCHH48 pKa = 6.12PGIHH52 pKa = 5.45RR53 pKa = 11.84TITFLQRR60 pKa = 11.84YY61 pKa = 7.1FWWPTLSRR69 pKa = 11.84DD70 pKa = 3.26AKK72 pKa = 10.04EE73 pKa = 4.01YY74 pKa = 11.13VSACTTCARR83 pKa = 11.84NKK85 pKa = 10.05IPNQPPSGSLHH96 pKa = 6.57PLPTPTRR103 pKa = 11.84PWSHH107 pKa = 5.83VSLDD111 pKa = 3.96FVTGLPPSAGNTVILTIIDD130 pKa = 3.92RR131 pKa = 11.84FSKK134 pKa = 10.35AAHH137 pKa = 5.77FVALPKK143 pKa = 10.68LPTALEE149 pKa = 4.09TARR152 pKa = 11.84LLTTHH157 pKa = 6.2VFRR160 pKa = 11.84LHH162 pKa = 7.38GIPDD166 pKa = 5.07DD167 pKa = 3.73IVSDD171 pKa = 4.23RR172 pKa = 11.84GPQFTSRR179 pKa = 11.84VWRR182 pKa = 11.84EE183 pKa = 3.45FCTSLGSKK191 pKa = 10.28VSLSSGYY198 pKa = 10.08HH199 pKa = 5.1PQSNGQSEE207 pKa = 4.5RR208 pKa = 11.84VNQEE212 pKa = 3.15LEE214 pKa = 3.68AALRR218 pKa = 11.84CVVSSNQAVWSEE230 pKa = 3.87EE231 pKa = 4.15LPWIEE236 pKa = 3.99YY237 pKa = 9.84AHH239 pKa = 6.68NSMTSSATGRR249 pKa = 11.84SPFEE253 pKa = 4.14ASLGFQPPLFPSIEE267 pKa = 4.32GEE269 pKa = 3.9HH270 pKa = 6.15SVPSVRR276 pKa = 11.84AHH278 pKa = 6.45LRR280 pKa = 11.84RR281 pKa = 11.84CRR283 pKa = 11.84RR284 pKa = 11.84VWKK287 pKa = 9.0ATRR290 pKa = 11.84AAFLRR295 pKa = 11.84TKK297 pKa = 10.55DD298 pKa = 3.37RR299 pKa = 11.84NKK301 pKa = 10.28RR302 pKa = 11.84FADD305 pKa = 3.8RR306 pKa = 11.84RR307 pKa = 11.84RR308 pKa = 11.84SSAPAYY314 pKa = 9.29TVGQKK319 pKa = 9.12VWLSTRR325 pKa = 11.84FVPIRR330 pKa = 11.84AEE332 pKa = 4.01SRR334 pKa = 11.84KK335 pKa = 10.02LSPAYY340 pKa = 9.42IGPFEE345 pKa = 4.67ISALVNN351 pKa = 3.57
WW1 pKa = 7.07GLSPEE6 pKa = 4.09KK7 pKa = 10.27SSRR10 pKa = 11.84PFARR14 pKa = 11.84LRR16 pKa = 11.84EE17 pKa = 4.06PNRR20 pKa = 11.84IQEE23 pKa = 4.26AALFVPQPVRR33 pKa = 11.84SQILQWAHH41 pKa = 5.04TARR44 pKa = 11.84FSCHH48 pKa = 6.12PGIHH52 pKa = 5.45RR53 pKa = 11.84TITFLQRR60 pKa = 11.84YY61 pKa = 7.1FWWPTLSRR69 pKa = 11.84DD70 pKa = 3.26AKK72 pKa = 10.04EE73 pKa = 4.01YY74 pKa = 11.13VSACTTCARR83 pKa = 11.84NKK85 pKa = 10.05IPNQPPSGSLHH96 pKa = 6.57PLPTPTRR103 pKa = 11.84PWSHH107 pKa = 5.83VSLDD111 pKa = 3.96FVTGLPPSAGNTVILTIIDD130 pKa = 3.92RR131 pKa = 11.84FSKK134 pKa = 10.35AAHH137 pKa = 5.77FVALPKK143 pKa = 10.68LPTALEE149 pKa = 4.09TARR152 pKa = 11.84LLTTHH157 pKa = 6.2VFRR160 pKa = 11.84LHH162 pKa = 7.38GIPDD166 pKa = 5.07DD167 pKa = 3.73IVSDD171 pKa = 4.23RR172 pKa = 11.84GPQFTSRR179 pKa = 11.84VWRR182 pKa = 11.84EE183 pKa = 3.45FCTSLGSKK191 pKa = 10.28VSLSSGYY198 pKa = 10.08HH199 pKa = 5.1PQSNGQSEE207 pKa = 4.5RR208 pKa = 11.84VNQEE212 pKa = 3.15LEE214 pKa = 3.68AALRR218 pKa = 11.84CVVSSNQAVWSEE230 pKa = 3.87EE231 pKa = 4.15LPWIEE236 pKa = 3.99YY237 pKa = 9.84AHH239 pKa = 6.68NSMTSSATGRR249 pKa = 11.84SPFEE253 pKa = 4.14ASLGFQPPLFPSIEE267 pKa = 4.32GEE269 pKa = 3.9HH270 pKa = 6.15SVPSVRR276 pKa = 11.84AHH278 pKa = 6.45LRR280 pKa = 11.84RR281 pKa = 11.84CRR283 pKa = 11.84RR284 pKa = 11.84VWKK287 pKa = 9.0ATRR290 pKa = 11.84AAFLRR295 pKa = 11.84TKK297 pKa = 10.55DD298 pKa = 3.37RR299 pKa = 11.84NKK301 pKa = 10.28RR302 pKa = 11.84FADD305 pKa = 3.8RR306 pKa = 11.84RR307 pKa = 11.84RR308 pKa = 11.84SSAPAYY314 pKa = 9.29TVGQKK319 pKa = 9.12VWLSTRR325 pKa = 11.84FVPIRR330 pKa = 11.84AEE332 pKa = 4.01SRR334 pKa = 11.84KK335 pKa = 10.02LSPAYY340 pKa = 9.42IGPFEE345 pKa = 4.67ISALVNN351 pKa = 3.57
Molecular weight: 39.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
19812588 |
17 |
8592 |
577.1 |
64.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.475 ± 0.01 | 2.23 ± 0.009 |
5.155 ± 0.009 | 6.863 ± 0.018 |
3.644 ± 0.01 | 6.238 ± 0.015 |
2.658 ± 0.007 | 4.468 ± 0.009 |
5.747 ± 0.012 | 9.466 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.364 ± 0.005 | 3.91 ± 0.007 |
5.716 ± 0.016 | 4.741 ± 0.011 |
5.555 ± 0.012 | 8.843 ± 0.017 |
5.696 ± 0.009 | 6.276 ± 0.01 |
1.138 ± 0.005 | 2.775 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
