
Nothophytophthora sp. Chile5
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Peronosporales; Peronosporaceae; Nothophytophthora; unclassified Nothophytophthora
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
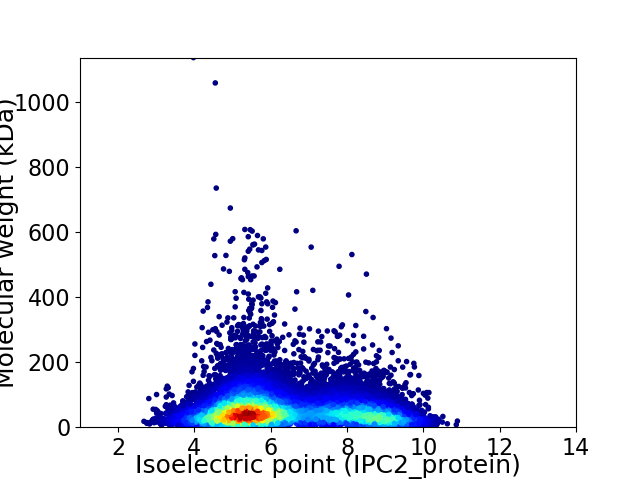
Virtual 2D-PAGE plot for 26905 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A662WEA1|A0A662WEA1_9STRA RING-type domain-containing protein OS=Nothophytophthora sp. Chile5 OX=2483409 GN=BBJ28_00020215 PE=4 SV=1
MM1 pKa = 7.41LSAAAFVSAAALLAASVDD19 pKa = 3.15AHH21 pKa = 7.35GYY23 pKa = 7.98MLIPEE28 pKa = 4.29SQFTGSANSAWIVQIDD44 pKa = 4.04PQWSSDD50 pKa = 2.97AWDD53 pKa = 4.26GNNQGSVDD61 pKa = 3.55AFVAAKK67 pKa = 9.88SANGYY72 pKa = 8.0TDD74 pKa = 5.06LRR76 pKa = 11.84TLMDD80 pKa = 3.78DD81 pKa = 3.46TSVYY85 pKa = 10.74GADD88 pKa = 4.3CGFTNPDD95 pKa = 2.78GTAQPVPTDD104 pKa = 3.38SKK106 pKa = 10.66ATFSRR111 pKa = 11.84ALVHH115 pKa = 6.79VGPCEE120 pKa = 3.49IWLDD124 pKa = 3.78DD125 pKa = 3.96TKK127 pKa = 11.53VLAEE131 pKa = 4.5DD132 pKa = 4.14DD133 pKa = 4.46CYY135 pKa = 11.69SLYY138 pKa = 11.16GNEE141 pKa = 4.22DD142 pKa = 3.1QDD144 pKa = 3.45IKK146 pKa = 11.41SIFPVDD152 pKa = 3.68YY153 pKa = 10.5SSCDD157 pKa = 3.33TAGCSMMRR165 pKa = 11.84FYY167 pKa = 10.87WLAFQGVDD175 pKa = 4.97GDD177 pKa = 4.86TVWQSYY183 pKa = 9.94KK184 pKa = 11.04DD185 pKa = 4.16CIPLEE190 pKa = 4.64GGSGTSSSTTTTDD203 pKa = 2.97APASEE208 pKa = 5.14ASATDD213 pKa = 3.62APSTDD218 pKa = 3.96APSTDD223 pKa = 4.13APATDD228 pKa = 4.8APTTDD233 pKa = 4.45APTTDD238 pKa = 4.45APTTDD243 pKa = 3.45APAAAEE249 pKa = 4.27TPEE252 pKa = 4.18VTPAPSDD259 pKa = 3.08KK260 pKa = 10.3CTGRR264 pKa = 11.84KK265 pKa = 8.85RR266 pKa = 11.84RR267 pKa = 11.84NN268 pKa = 3.03
MM1 pKa = 7.41LSAAAFVSAAALLAASVDD19 pKa = 3.15AHH21 pKa = 7.35GYY23 pKa = 7.98MLIPEE28 pKa = 4.29SQFTGSANSAWIVQIDD44 pKa = 4.04PQWSSDD50 pKa = 2.97AWDD53 pKa = 4.26GNNQGSVDD61 pKa = 3.55AFVAAKK67 pKa = 9.88SANGYY72 pKa = 8.0TDD74 pKa = 5.06LRR76 pKa = 11.84TLMDD80 pKa = 3.78DD81 pKa = 3.46TSVYY85 pKa = 10.74GADD88 pKa = 4.3CGFTNPDD95 pKa = 2.78GTAQPVPTDD104 pKa = 3.38SKK106 pKa = 10.66ATFSRR111 pKa = 11.84ALVHH115 pKa = 6.79VGPCEE120 pKa = 3.49IWLDD124 pKa = 3.78DD125 pKa = 3.96TKK127 pKa = 11.53VLAEE131 pKa = 4.5DD132 pKa = 4.14DD133 pKa = 4.46CYY135 pKa = 11.69SLYY138 pKa = 11.16GNEE141 pKa = 4.22DD142 pKa = 3.1QDD144 pKa = 3.45IKK146 pKa = 11.41SIFPVDD152 pKa = 3.68YY153 pKa = 10.5SSCDD157 pKa = 3.33TAGCSMMRR165 pKa = 11.84FYY167 pKa = 10.87WLAFQGVDD175 pKa = 4.97GDD177 pKa = 4.86TVWQSYY183 pKa = 9.94KK184 pKa = 11.04DD185 pKa = 4.16CIPLEE190 pKa = 4.64GGSGTSSSTTTTDD203 pKa = 2.97APASEE208 pKa = 5.14ASATDD213 pKa = 3.62APSTDD218 pKa = 3.96APSTDD223 pKa = 4.13APATDD228 pKa = 4.8APTTDD233 pKa = 4.45APTTDD238 pKa = 4.45APTTDD243 pKa = 3.45APAAAEE249 pKa = 4.27TPEE252 pKa = 4.18VTPAPSDD259 pKa = 3.08KK260 pKa = 10.3CTGRR264 pKa = 11.84KK265 pKa = 8.85RR266 pKa = 11.84RR267 pKa = 11.84NN268 pKa = 3.03
Molecular weight: 28.02 kDa
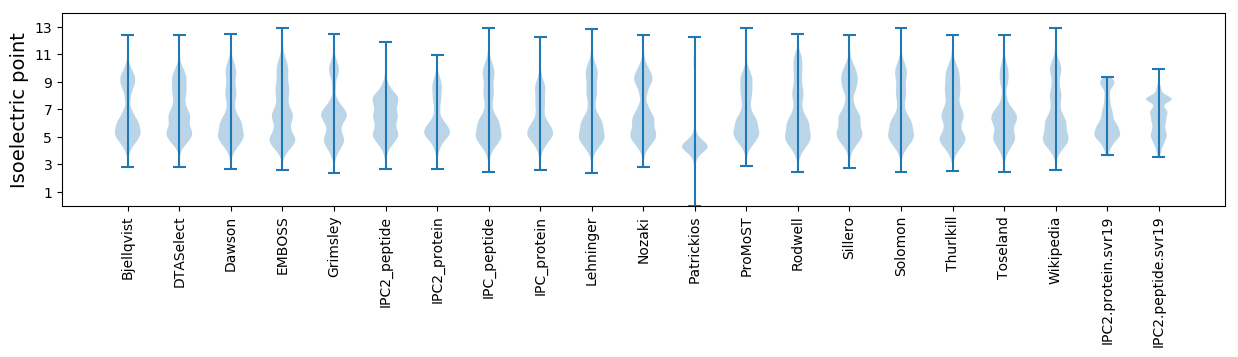
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A662X0T5|A0A662X0T5_9STRA Uncharacterized protein OS=Nothophytophthora sp. Chile5 OX=2483409 GN=BBJ28_00023662 PE=4 SV=1
MM1 pKa = 7.91PSNGCRR7 pKa = 11.84ISCILATIEE16 pKa = 5.31DD17 pKa = 3.9EE18 pKa = 4.52DD19 pKa = 3.9EE20 pKa = 4.16RR21 pKa = 11.84VATGVMTAFLSGVILLLVRR40 pKa = 11.84ILNLRR45 pKa = 11.84QVTDD49 pKa = 4.29FFSRR53 pKa = 11.84PVMDD57 pKa = 4.4GFISAGGLLIMTCSKK72 pKa = 10.7SRR74 pKa = 11.84WATRR78 pKa = 11.84HH79 pKa = 3.52TRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84MFQFFKK89 pKa = 10.75HH90 pKa = 5.72VGEE93 pKa = 4.66SKK95 pKa = 10.31PAALGLFKK103 pKa = 10.63KK104 pKa = 10.46RR105 pKa = 11.84PQCATRR111 pKa = 11.84HH112 pKa = 5.33FPRR115 pKa = 11.84THH117 pKa = 6.06HH118 pKa = 6.1VLPGLLVVCIGGGIAGYY135 pKa = 10.64LLGPKK140 pKa = 9.53ALKK143 pKa = 10.22LAANVPGGFPPPDD156 pKa = 3.74APWYY160 pKa = 9.96GFTSGLIVGGTILLNASTVSLVVFLSSIAMTKK192 pKa = 10.15RR193 pKa = 11.84LAIQRR198 pKa = 11.84GEE200 pKa = 4.47DD201 pKa = 3.14INTKK205 pKa = 10.0QEE207 pKa = 4.01LTGVGCASIVCGFFRR222 pKa = 11.84TMRR225 pKa = 11.84PTGGVSRR232 pKa = 11.84TAVNLQNAHH241 pKa = 5.6IQVPRR246 pKa = 11.84RR247 pKa = 11.84SLFSPRR253 pKa = 11.84CTFRR257 pKa = 11.84GTLYY261 pKa = 10.05YY262 pKa = 10.53LPRR265 pKa = 11.84AALASIIIATSYY277 pKa = 11.07SLPPMM282 pKa = 4.55
MM1 pKa = 7.91PSNGCRR7 pKa = 11.84ISCILATIEE16 pKa = 5.31DD17 pKa = 3.9EE18 pKa = 4.52DD19 pKa = 3.9EE20 pKa = 4.16RR21 pKa = 11.84VATGVMTAFLSGVILLLVRR40 pKa = 11.84ILNLRR45 pKa = 11.84QVTDD49 pKa = 4.29FFSRR53 pKa = 11.84PVMDD57 pKa = 4.4GFISAGGLLIMTCSKK72 pKa = 10.7SRR74 pKa = 11.84WATRR78 pKa = 11.84HH79 pKa = 3.52TRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84MFQFFKK89 pKa = 10.75HH90 pKa = 5.72VGEE93 pKa = 4.66SKK95 pKa = 10.31PAALGLFKK103 pKa = 10.63KK104 pKa = 10.46RR105 pKa = 11.84PQCATRR111 pKa = 11.84HH112 pKa = 5.33FPRR115 pKa = 11.84THH117 pKa = 6.06HH118 pKa = 6.1VLPGLLVVCIGGGIAGYY135 pKa = 10.64LLGPKK140 pKa = 9.53ALKK143 pKa = 10.22LAANVPGGFPPPDD156 pKa = 3.74APWYY160 pKa = 9.96GFTSGLIVGGTILLNASTVSLVVFLSSIAMTKK192 pKa = 10.15RR193 pKa = 11.84LAIQRR198 pKa = 11.84GEE200 pKa = 4.47DD201 pKa = 3.14INTKK205 pKa = 10.0QEE207 pKa = 4.01LTGVGCASIVCGFFRR222 pKa = 11.84TMRR225 pKa = 11.84PTGGVSRR232 pKa = 11.84TAVNLQNAHH241 pKa = 5.6IQVPRR246 pKa = 11.84RR247 pKa = 11.84SLFSPRR253 pKa = 11.84CTFRR257 pKa = 11.84GTLYY261 pKa = 10.05YY262 pKa = 10.53LPRR265 pKa = 11.84AALASIIIATSYY277 pKa = 11.07SLPPMM282 pKa = 4.55
Molecular weight: 30.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13882063 |
49 |
10584 |
516.0 |
56.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.217 ± 0.018 | 1.607 ± 0.006 |
5.597 ± 0.01 | 6.691 ± 0.015 |
3.696 ± 0.009 | 6.376 ± 0.014 |
2.327 ± 0.006 | 3.544 ± 0.011 |
4.439 ± 0.015 | 9.808 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.306 ± 0.007 | 3.026 ± 0.007 |
4.853 ± 0.012 | 4.33 ± 0.01 |
6.427 ± 0.016 | 8.197 ± 0.015 |
5.751 ± 0.012 | 7.231 ± 0.011 |
1.195 ± 0.005 | 2.382 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
