
Hydrogenivirga caldilitoris
Taxonomy: cellular organisms; Bacteria; Aquificae; Aquificae; Aquificales; Aquificaceae; Hydrogenivirga
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
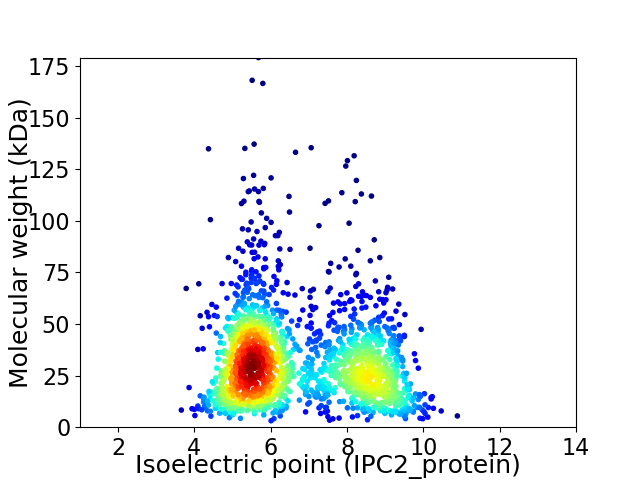
Virtual 2D-PAGE plot for 1885 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A497XMM9|A0A497XMM9_9AQUI Uncharacterized protein OS=Hydrogenivirga caldilitoris OX=246264 GN=BCF55_0409 PE=4 SV=1
MM1 pKa = 7.48SKK3 pKa = 10.59KK4 pKa = 10.08FLLSAVFSAGVLSAGALAGDD24 pKa = 3.4MQVLNEE30 pKa = 4.47EE31 pKa = 4.16EE32 pKa = 4.74LEE34 pKa = 4.23DD35 pKa = 4.33VSAQGLQVIEE45 pKa = 3.8NDD47 pKa = 2.75NRR49 pKa = 11.84QFAEE53 pKa = 4.48INGQNNNLDD62 pKa = 3.89SVQLNDD68 pKa = 3.39NAQLEE73 pKa = 4.36AQAGGIINVAKK84 pKa = 9.32TALNAAVNLLADD96 pKa = 4.38GPEE99 pKa = 4.65PGPKK103 pKa = 9.65PPPQNGDD110 pKa = 2.94TKK112 pKa = 10.72YY113 pKa = 9.82FNYY116 pKa = 10.3YY117 pKa = 10.27FSQTNVQVAMNHH129 pKa = 5.24VNTANNADD137 pKa = 3.92AEE139 pKa = 4.46EE140 pKa = 4.16EE141 pKa = 3.97LAIAGNLNKK150 pKa = 8.79EE151 pKa = 4.44TQSIDD156 pKa = 3.1NNGNIRR162 pKa = 11.84EE163 pKa = 3.93QDD165 pKa = 3.38NNNNSVQLNDD175 pKa = 3.59SAQSGASGLMIEE187 pKa = 4.69NAAISAVNTGMNIFVAGDD205 pKa = 3.29VDD207 pKa = 4.08STEE210 pKa = 4.04GHH212 pKa = 4.42QTNTQVAKK220 pKa = 11.21NMGNYY225 pKa = 10.22AEE227 pKa = 4.86AQAGDD232 pKa = 3.66QPIAVAGNAEE242 pKa = 4.39LGVTQNINNQYY253 pKa = 9.9QSEE256 pKa = 4.47GEE258 pKa = 4.04PNNIYY263 pKa = 10.88DD264 pKa = 4.25QDD266 pKa = 3.9NNNNSVQLNDD276 pKa = 3.59SAQSGASAVSMLNVANSAYY295 pKa = 10.59NSGLNVMAMANLSNSLMTQNNSQTAEE321 pKa = 3.84SHH323 pKa = 5.94VNVASAEE330 pKa = 4.03ADD332 pKa = 3.29NGVALAGNLNKK343 pKa = 8.77QTQNVHH349 pKa = 5.81NAVADD354 pKa = 3.98DD355 pKa = 4.04FVGLGVIEE363 pKa = 4.32EE364 pKa = 4.07QDD366 pKa = 3.43NNNNSVQLNDD376 pKa = 3.59SAQSGASALDD386 pKa = 3.58MTNSAVSAGNTGMNVMAATTVSTSEE411 pKa = 3.72IHH413 pKa = 5.7QGNYY417 pKa = 7.66QSARR421 pKa = 11.84NFDD424 pKa = 3.72NEE426 pKa = 3.32ARR428 pKa = 11.84TYY430 pKa = 10.73NGNPEE435 pKa = 4.13SEE437 pKa = 5.19DD438 pKa = 3.03IAVATNAEE446 pKa = 4.24IAGEE450 pKa = 4.0PGQYY454 pKa = 9.75VHH456 pKa = 6.66TVHH459 pKa = 6.65ATIGDD464 pKa = 3.68QNNNNNSVQLNDD476 pKa = 4.37DD477 pKa = 3.98AQSSASALFMTNMAQSAVNSGLNLLFVDD505 pKa = 4.95GNVDD509 pKa = 3.81DD510 pKa = 4.54STVSQTNEE518 pKa = 3.54QFASNHH524 pKa = 5.12NNYY527 pKa = 8.3ATANALAVAINLNKK541 pKa = 9.49QRR543 pKa = 11.84QIIEE547 pKa = 3.95NCFCSDD553 pKa = 5.23LSQTDD558 pKa = 2.95QDD560 pKa = 3.83NNMNSVQLNDD570 pKa = 3.33NAQSYY575 pKa = 10.21ASGWMIVNAATSAVNSGTNLAVVNGVVSGSSVTQTNVQTSVNFSNQASGNTAIAGNAEE633 pKa = 4.14IGSLFIPIGFF643 pKa = 3.97
MM1 pKa = 7.48SKK3 pKa = 10.59KK4 pKa = 10.08FLLSAVFSAGVLSAGALAGDD24 pKa = 3.4MQVLNEE30 pKa = 4.47EE31 pKa = 4.16EE32 pKa = 4.74LEE34 pKa = 4.23DD35 pKa = 4.33VSAQGLQVIEE45 pKa = 3.8NDD47 pKa = 2.75NRR49 pKa = 11.84QFAEE53 pKa = 4.48INGQNNNLDD62 pKa = 3.89SVQLNDD68 pKa = 3.39NAQLEE73 pKa = 4.36AQAGGIINVAKK84 pKa = 9.32TALNAAVNLLADD96 pKa = 4.38GPEE99 pKa = 4.65PGPKK103 pKa = 9.65PPPQNGDD110 pKa = 2.94TKK112 pKa = 10.72YY113 pKa = 9.82FNYY116 pKa = 10.3YY117 pKa = 10.27FSQTNVQVAMNHH129 pKa = 5.24VNTANNADD137 pKa = 3.92AEE139 pKa = 4.46EE140 pKa = 4.16EE141 pKa = 3.97LAIAGNLNKK150 pKa = 8.79EE151 pKa = 4.44TQSIDD156 pKa = 3.1NNGNIRR162 pKa = 11.84EE163 pKa = 3.93QDD165 pKa = 3.38NNNNSVQLNDD175 pKa = 3.59SAQSGASGLMIEE187 pKa = 4.69NAAISAVNTGMNIFVAGDD205 pKa = 3.29VDD207 pKa = 4.08STEE210 pKa = 4.04GHH212 pKa = 4.42QTNTQVAKK220 pKa = 11.21NMGNYY225 pKa = 10.22AEE227 pKa = 4.86AQAGDD232 pKa = 3.66QPIAVAGNAEE242 pKa = 4.39LGVTQNINNQYY253 pKa = 9.9QSEE256 pKa = 4.47GEE258 pKa = 4.04PNNIYY263 pKa = 10.88DD264 pKa = 4.25QDD266 pKa = 3.9NNNNSVQLNDD276 pKa = 3.59SAQSGASAVSMLNVANSAYY295 pKa = 10.59NSGLNVMAMANLSNSLMTQNNSQTAEE321 pKa = 3.84SHH323 pKa = 5.94VNVASAEE330 pKa = 4.03ADD332 pKa = 3.29NGVALAGNLNKK343 pKa = 8.77QTQNVHH349 pKa = 5.81NAVADD354 pKa = 3.98DD355 pKa = 4.04FVGLGVIEE363 pKa = 4.32EE364 pKa = 4.07QDD366 pKa = 3.43NNNNSVQLNDD376 pKa = 3.59SAQSGASALDD386 pKa = 3.58MTNSAVSAGNTGMNVMAATTVSTSEE411 pKa = 3.72IHH413 pKa = 5.7QGNYY417 pKa = 7.66QSARR421 pKa = 11.84NFDD424 pKa = 3.72NEE426 pKa = 3.32ARR428 pKa = 11.84TYY430 pKa = 10.73NGNPEE435 pKa = 4.13SEE437 pKa = 5.19DD438 pKa = 3.03IAVATNAEE446 pKa = 4.24IAGEE450 pKa = 4.0PGQYY454 pKa = 9.75VHH456 pKa = 6.66TVHH459 pKa = 6.65ATIGDD464 pKa = 3.68QNNNNNSVQLNDD476 pKa = 4.37DD477 pKa = 3.98AQSSASALFMTNMAQSAVNSGLNLLFVDD505 pKa = 4.95GNVDD509 pKa = 3.81DD510 pKa = 4.54STVSQTNEE518 pKa = 3.54QFASNHH524 pKa = 5.12NNYY527 pKa = 8.3ATANALAVAINLNKK541 pKa = 9.49QRR543 pKa = 11.84QIIEE547 pKa = 3.95NCFCSDD553 pKa = 5.23LSQTDD558 pKa = 2.95QDD560 pKa = 3.83NNMNSVQLNDD570 pKa = 3.33NAQSYY575 pKa = 10.21ASGWMIVNAATSAVNSGTNLAVVNGVVSGSSVTQTNVQTSVNFSNQASGNTAIAGNAEE633 pKa = 4.14IGSLFIPIGFF643 pKa = 3.97
Molecular weight: 67.23 kDa
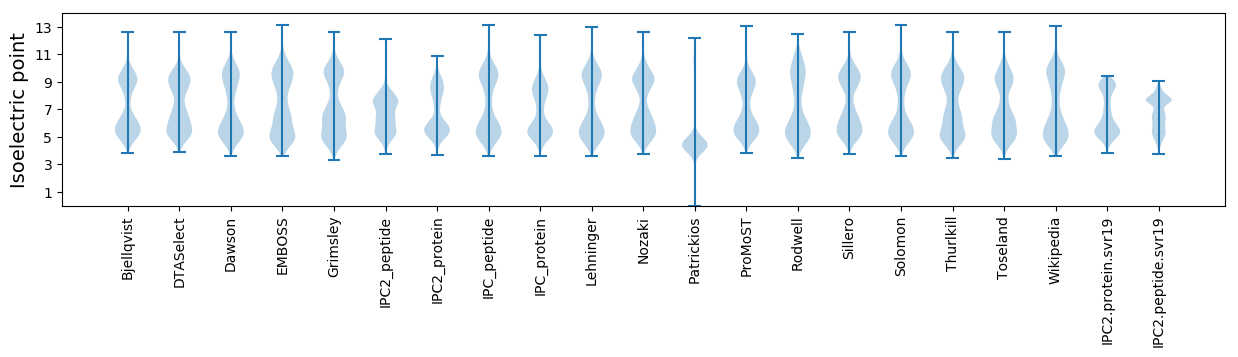
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A497XMI3|A0A497XMI3_9AQUI Prepilin-type N-terminal cleavage/methylation domain-containing protein OS=Hydrogenivirga caldilitoris OX=246264 GN=BCF55_0390 PE=4 SV=1
MM1 pKa = 7.76PEE3 pKa = 3.81ALKK6 pKa = 10.3PHH8 pKa = 6.36ISNRR12 pKa = 11.84KK13 pKa = 8.03KK14 pKa = 10.01KK15 pKa = 10.01RR16 pKa = 11.84KK17 pKa = 9.55SGFLARR23 pKa = 11.84MKK25 pKa = 10.55SRR27 pKa = 11.84AGRR30 pKa = 11.84KK31 pKa = 8.0VIKK34 pKa = 10.02RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84QKK39 pKa = 10.21GRR41 pKa = 11.84KK42 pKa = 8.77RR43 pKa = 11.84LAPP46 pKa = 3.99
MM1 pKa = 7.76PEE3 pKa = 3.81ALKK6 pKa = 10.3PHH8 pKa = 6.36ISNRR12 pKa = 11.84KK13 pKa = 8.03KK14 pKa = 10.01KK15 pKa = 10.01RR16 pKa = 11.84KK17 pKa = 9.55SGFLARR23 pKa = 11.84MKK25 pKa = 10.55SRR27 pKa = 11.84AGRR30 pKa = 11.84KK31 pKa = 8.0VIKK34 pKa = 10.02RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84QKK39 pKa = 10.21GRR41 pKa = 11.84KK42 pKa = 8.77RR43 pKa = 11.84LAPP46 pKa = 3.99
Molecular weight: 5.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
561480 |
30 |
1582 |
297.9 |
33.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.222 ± 0.052 | 0.837 ± 0.023 |
4.67 ± 0.041 | 9.048 ± 0.082 |
4.915 ± 0.054 | 7.314 ± 0.048 |
1.575 ± 0.021 | 6.823 ± 0.049 |
7.407 ± 0.066 | 10.973 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.091 ± 0.022 | 3.294 ± 0.033 |
4.098 ± 0.037 | 2.114 ± 0.025 |
5.879 ± 0.05 | 5.443 ± 0.046 |
4.394 ± 0.034 | 8.15 ± 0.04 |
0.958 ± 0.021 | 3.796 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
