
Varibaculum cambriense DORA_20
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Varibaculum; Varibaculum cambriense
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
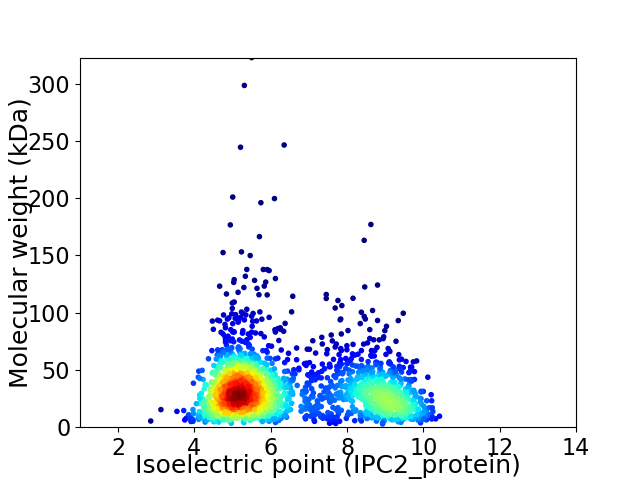
Virtual 2D-PAGE plot for 2003 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
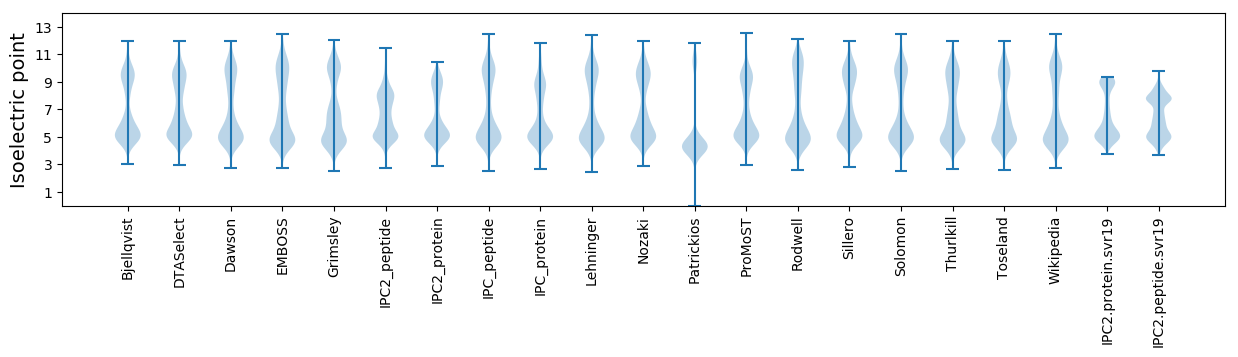
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W1TWR6|W1TWR6_9ACTO PKS_ER domain-containing protein OS=Varibaculum cambriense DORA_20 OX=1403948 GN=Q618_VCMC00001G1334 PE=4 SV=1
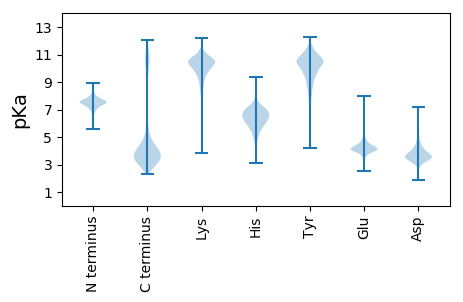
MM1 pKa = 7.66TYY3 pKa = 10.9DD4 pKa = 3.46EE5 pKa = 5.75LYY7 pKa = 7.87VTSRR11 pKa = 11.84AFFEE15 pKa = 4.33AHH17 pKa = 6.02ATPEE21 pKa = 3.95QIAFLDD27 pKa = 3.73SHH29 pKa = 6.35EE30 pKa = 4.67KK31 pKa = 10.42CDD33 pKa = 3.54GAGFVDD39 pKa = 5.14SILFFIDD46 pKa = 4.76LFLGDD51 pKa = 3.98MPGVPEE57 pKa = 5.99KK58 pKa = 10.56LANWQASLDD67 pKa = 3.82ALPEE71 pKa = 3.85YY72 pKa = 10.64DD73 pKa = 3.64
MM1 pKa = 7.66TYY3 pKa = 10.9DD4 pKa = 3.46EE5 pKa = 5.75LYY7 pKa = 7.87VTSRR11 pKa = 11.84AFFEE15 pKa = 4.33AHH17 pKa = 6.02ATPEE21 pKa = 3.95QIAFLDD27 pKa = 3.73SHH29 pKa = 6.35EE30 pKa = 4.67KK31 pKa = 10.42CDD33 pKa = 3.54GAGFVDD39 pKa = 5.14SILFFIDD46 pKa = 4.76LFLGDD51 pKa = 3.98MPGVPEE57 pKa = 5.99KK58 pKa = 10.56LANWQASLDD67 pKa = 3.82ALPEE71 pKa = 3.85YY72 pKa = 10.64DD73 pKa = 3.64
Molecular weight: 8.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W1TTC0|W1TTC0_9ACTO Probable nicotinate-nucleotide adenylyltransferase OS=Varibaculum cambriense DORA_20 OX=1403948 GN=nadD PE=3 SV=1
MM1 pKa = 7.4SRR3 pKa = 11.84TFTSFKK9 pKa = 8.25YY10 pKa = 10.34HH11 pKa = 7.13NYY13 pKa = 9.06RR14 pKa = 11.84LWWMSNIFASTATWMQRR31 pKa = 11.84VAQDD35 pKa = 3.21WVVLMVLTDD44 pKa = 3.3QSGFAVGVVTALQFLPQLLLSIPAGMVADD73 pKa = 4.45RR74 pKa = 11.84FNRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84IIQICQCIVALCGLLTGILLLTGVAALWHH108 pKa = 6.62IYY110 pKa = 9.81IIALITGVADD120 pKa = 4.1CLTSPARR127 pKa = 11.84QAFVSEE133 pKa = 4.57LVTKK137 pKa = 10.45EE138 pKa = 4.19DD139 pKa = 3.49LPNAVGLNSTAFNSARR155 pKa = 11.84LIGPGLAGFVIAGIGPGWVFIANFVLFIVPVLGLSLMDD193 pKa = 3.94PARR196 pKa = 11.84LYY198 pKa = 10.82RR199 pKa = 11.84PEE201 pKa = 3.83KK202 pKa = 8.86TSRR205 pKa = 11.84TKK207 pKa = 11.1GMMRR211 pKa = 11.84EE212 pKa = 3.97GLRR215 pKa = 11.84YY216 pKa = 8.65VQNRR220 pKa = 11.84TDD222 pKa = 2.95IKK224 pKa = 10.94AIIAILTVVGALGLNFQLTQAVMATRR250 pKa = 11.84VFGKK254 pKa = 10.62GAGEE258 pKa = 3.92YY259 pKa = 10.51GLLGSIMAIGALAGALQAARR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84QPRR284 pKa = 11.84VFTVVVSAVLFGVLEE299 pKa = 4.4GALALAPTYY308 pKa = 10.36EE309 pKa = 4.09IFALLLIPTGFVMLNLLTSANATIQVSTEE338 pKa = 3.55EE339 pKa = 4.32EE340 pKa = 3.49IRR342 pKa = 11.84GRR344 pKa = 11.84VLSIYY349 pKa = 10.44FLFFLGTTPIGAPIIGWVAEE369 pKa = 3.96HH370 pKa = 6.94WGPRR374 pKa = 11.84WSLGAGAIASLLVALIVGIWLWRR397 pKa = 11.84HH398 pKa = 4.39WKK400 pKa = 10.32VDD402 pKa = 3.28VTLRR406 pKa = 11.84ASRR409 pKa = 11.84PFVSIEE415 pKa = 3.96GPRR418 pKa = 11.84EE419 pKa = 3.18RR420 pKa = 11.84AMRR423 pKa = 11.84RR424 pKa = 11.84RR425 pKa = 11.84KK426 pKa = 9.37EE427 pKa = 3.49RR428 pKa = 11.84AEE430 pKa = 3.9RR431 pKa = 11.84QRR433 pKa = 11.84IMDD436 pKa = 3.72NQSRR440 pKa = 11.84DD441 pKa = 3.12TGTT444 pKa = 3.0
MM1 pKa = 7.4SRR3 pKa = 11.84TFTSFKK9 pKa = 8.25YY10 pKa = 10.34HH11 pKa = 7.13NYY13 pKa = 9.06RR14 pKa = 11.84LWWMSNIFASTATWMQRR31 pKa = 11.84VAQDD35 pKa = 3.21WVVLMVLTDD44 pKa = 3.3QSGFAVGVVTALQFLPQLLLSIPAGMVADD73 pKa = 4.45RR74 pKa = 11.84FNRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84IIQICQCIVALCGLLTGILLLTGVAALWHH108 pKa = 6.62IYY110 pKa = 9.81IIALITGVADD120 pKa = 4.1CLTSPARR127 pKa = 11.84QAFVSEE133 pKa = 4.57LVTKK137 pKa = 10.45EE138 pKa = 4.19DD139 pKa = 3.49LPNAVGLNSTAFNSARR155 pKa = 11.84LIGPGLAGFVIAGIGPGWVFIANFVLFIVPVLGLSLMDD193 pKa = 3.94PARR196 pKa = 11.84LYY198 pKa = 10.82RR199 pKa = 11.84PEE201 pKa = 3.83KK202 pKa = 8.86TSRR205 pKa = 11.84TKK207 pKa = 11.1GMMRR211 pKa = 11.84EE212 pKa = 3.97GLRR215 pKa = 11.84YY216 pKa = 8.65VQNRR220 pKa = 11.84TDD222 pKa = 2.95IKK224 pKa = 10.94AIIAILTVVGALGLNFQLTQAVMATRR250 pKa = 11.84VFGKK254 pKa = 10.62GAGEE258 pKa = 3.92YY259 pKa = 10.51GLLGSIMAIGALAGALQAARR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84QPRR284 pKa = 11.84VFTVVVSAVLFGVLEE299 pKa = 4.4GALALAPTYY308 pKa = 10.36EE309 pKa = 4.09IFALLLIPTGFVMLNLLTSANATIQVSTEE338 pKa = 3.55EE339 pKa = 4.32EE340 pKa = 3.49IRR342 pKa = 11.84GRR344 pKa = 11.84VLSIYY349 pKa = 10.44FLFFLGTTPIGAPIIGWVAEE369 pKa = 3.96HH370 pKa = 6.94WGPRR374 pKa = 11.84WSLGAGAIASLLVALIVGIWLWRR397 pKa = 11.84HH398 pKa = 4.39WKK400 pKa = 10.32VDD402 pKa = 3.28VTLRR406 pKa = 11.84ASRR409 pKa = 11.84PFVSIEE415 pKa = 3.96GPRR418 pKa = 11.84EE419 pKa = 3.18RR420 pKa = 11.84AMRR423 pKa = 11.84RR424 pKa = 11.84RR425 pKa = 11.84KK426 pKa = 9.37EE427 pKa = 3.49RR428 pKa = 11.84AEE430 pKa = 3.9RR431 pKa = 11.84QRR433 pKa = 11.84IMDD436 pKa = 3.72NQSRR440 pKa = 11.84DD441 pKa = 3.12TGTT444 pKa = 3.0
Molecular weight: 48.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
657404 |
26 |
2987 |
328.2 |
35.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.841 ± 0.066 | 0.956 ± 0.02 |
5.376 ± 0.043 | 6.106 ± 0.06 |
3.249 ± 0.034 | 8.017 ± 0.047 |
1.787 ± 0.026 | 5.324 ± 0.047 |
4.578 ± 0.049 | 9.785 ± 0.075 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.16 ± 0.03 | 3.181 ± 0.039 |
5.03 ± 0.04 | 4.203 ± 0.036 |
5.82 ± 0.056 | 6.365 ± 0.045 |
5.745 ± 0.051 | 7.373 ± 0.045 |
1.385 ± 0.027 | 2.528 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
