
Desulfocurvibacter africanus subsp. africanus str. Walvis Bay
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfocurvibacter; Desulfocurvibacter africanus; Desulfocurvibacter africanus subsp. africanus
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
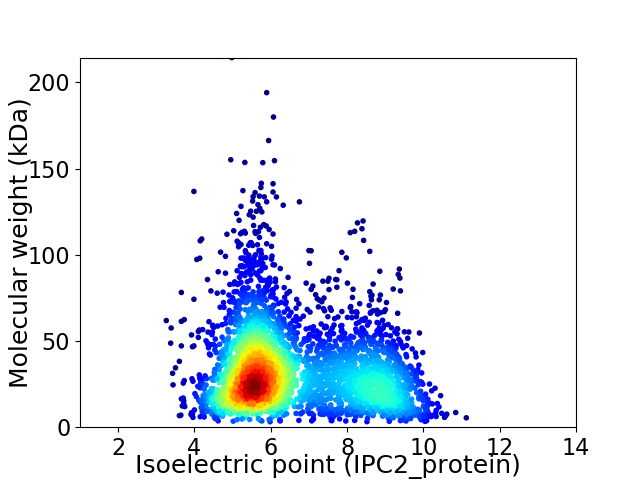
Virtual 2D-PAGE plot for 3699 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F3Z1C3|F3Z1C3_DESAF Uncharacterized protein OS=Desulfocurvibacter africanus subsp. africanus str. Walvis Bay OX=690850 GN=Desaf_2813 PE=4 SV=1
MM1 pKa = 7.78SDD3 pKa = 3.05AKK5 pKa = 10.79SAIGTVVKK13 pKa = 10.74ISGQVFAEE21 pKa = 4.34ADD23 pKa = 3.64GHH25 pKa = 5.09RR26 pKa = 11.84HH27 pKa = 5.53PLAEE31 pKa = 4.33GSTVRR36 pKa = 11.84EE37 pKa = 4.34DD38 pKa = 3.66EE39 pKa = 4.55TVVTADD45 pKa = 3.26GSKK48 pKa = 10.32VEE50 pKa = 5.59IRR52 pKa = 11.84FTDD55 pKa = 3.49NTVLSQGEE63 pKa = 3.95NSRR66 pKa = 11.84IEE68 pKa = 3.92LDD70 pKa = 3.44EE71 pKa = 3.89YY72 pKa = 11.43VYY74 pKa = 11.1EE75 pKa = 4.44GDD77 pKa = 3.83KK78 pKa = 11.18GAAGLLFNMVQGSFRR93 pKa = 11.84TVTGKK98 pKa = 9.85IVEE101 pKa = 4.18QNPEE105 pKa = 4.07GFNLSSPLATIGIRR119 pKa = 11.84GTTTFHH125 pKa = 6.99QIGPDD130 pKa = 3.85GEE132 pKa = 4.22RR133 pKa = 11.84HH134 pKa = 4.88GVQNIANPSLGGLDD148 pKa = 3.82SPLHH152 pKa = 5.57GMPSGYY158 pKa = 8.48HH159 pKa = 4.76TMLVTFSNGEE169 pKa = 3.89TRR171 pKa = 11.84VIADD175 pKa = 4.2SYY177 pKa = 11.67QMVDD181 pKa = 3.46LTSAGIGMIRR191 pKa = 11.84TFSLNEE197 pKa = 3.78LLDD200 pKa = 4.87FIDD203 pKa = 5.33LSAHH207 pKa = 5.28SPEE210 pKa = 4.15EE211 pKa = 4.09RR212 pKa = 11.84EE213 pKa = 4.32SLVDD217 pKa = 3.51TLLHH221 pKa = 6.16EE222 pKa = 5.0FRR224 pKa = 11.84EE225 pKa = 4.39QVPPEE230 pKa = 4.25PQSEE234 pKa = 4.42HH235 pKa = 5.94QSSSGDD241 pKa = 3.44NLSPDD246 pKa = 3.35VTDD249 pKa = 6.37DD250 pKa = 3.47GTQDD254 pKa = 3.91SDD256 pKa = 5.74GEE258 pKa = 4.14DD259 pKa = 3.09QQAQDD264 pKa = 4.45EE265 pKa = 4.8VPPDD269 pKa = 3.39IQRR272 pKa = 11.84IISDD276 pKa = 4.32PLPEE280 pKa = 4.14PVIQGGGGNGGGWNPPQEE298 pKa = 4.77DD299 pKa = 4.11PPVRR303 pKa = 11.84DD304 pKa = 3.67VIEE307 pKa = 4.63IIPRR311 pKa = 11.84ITFPIFLIGDD321 pKa = 4.28GGNNSIEE328 pKa = 4.06GTEE331 pKa = 4.13YY332 pKa = 10.39PDD334 pKa = 5.61FIQGLAGDD342 pKa = 4.18DD343 pKa = 4.09SLWGLGSNDD352 pKa = 3.38TLQGGEE358 pKa = 4.49GNDD361 pKa = 3.53VLYY364 pKa = 11.01GGAGDD369 pKa = 4.75DD370 pKa = 3.77VLDD373 pKa = 4.5GGRR376 pKa = 11.84GDD378 pKa = 3.68NSIDD382 pKa = 3.6GGAGNNWVSYY392 pKa = 10.98ASVRR396 pKa = 11.84DD397 pKa = 3.6TFQNKK402 pKa = 8.8GVVVYY407 pKa = 10.81LDD409 pKa = 4.61DD410 pKa = 5.39EE411 pKa = 6.25GDD413 pKa = 3.31DD414 pKa = 3.89SYY416 pKa = 12.02GVRR419 pKa = 11.84EE420 pKa = 4.08YY421 pKa = 10.35PVYY424 pKa = 10.44EE425 pKa = 4.29SNGNLLSEE433 pKa = 4.29PHH435 pKa = 6.3SEE437 pKa = 4.05YY438 pKa = 11.2DD439 pKa = 3.91DD440 pKa = 4.2LASIANVEE448 pKa = 4.05GSIFSDD454 pKa = 4.13AIYY457 pKa = 10.83GNDD460 pKa = 4.01HH461 pKa = 6.43NNHH464 pKa = 6.17LKK466 pKa = 10.72GLAGDD471 pKa = 4.39DD472 pKa = 4.32SIIGNAGDD480 pKa = 3.73DD481 pKa = 4.12TLDD484 pKa = 3.79GGQGNDD490 pKa = 4.06TIDD493 pKa = 3.64GGLGSDD499 pKa = 4.12VIIGGSGNDD508 pKa = 3.06WVSYY512 pKa = 9.35QSYY515 pKa = 9.69YY516 pKa = 10.63SCSFQGMSIDD526 pKa = 4.38LVNGACSLDD535 pKa = 3.63TSSEE539 pKa = 4.06DD540 pKa = 3.25TDD542 pKa = 3.45QFSGIEE548 pKa = 3.95NVEE551 pKa = 3.72GSEE554 pKa = 4.27YY555 pKa = 10.92GDD557 pKa = 4.36EE558 pKa = 4.19ICGDD562 pKa = 3.65SLGNTLHH569 pKa = 6.79GMGGSDD575 pKa = 4.27TISGGAGSDD584 pKa = 3.95FINGGEE590 pKa = 4.97GIDD593 pKa = 3.6TLYY596 pKa = 11.46GNAGADD602 pKa = 3.08TFYY605 pKa = 10.75FDD607 pKa = 4.37NMEE610 pKa = 5.12DD611 pKa = 3.63GFSVDD616 pKa = 2.93THH618 pKa = 8.34ISFSGQQGDD627 pKa = 4.3LIKK630 pKa = 10.95DD631 pKa = 3.79FTPGQDD637 pKa = 4.05KK638 pKa = 10.81ILLDD642 pKa = 3.71SSQYY646 pKa = 9.65SQLGMVVSYY655 pKa = 7.96DD656 pKa = 3.62TNFITIDD663 pKa = 3.37GLYY666 pKa = 10.45DD667 pKa = 3.47GNNATDD673 pKa = 3.37VQGAYY678 pKa = 10.46SSWLNGEE685 pKa = 3.68ACLIVDD691 pKa = 3.76RR692 pKa = 11.84DD693 pKa = 4.01YY694 pKa = 12.18VNDD697 pKa = 4.04CCRR700 pKa = 11.84LIYY703 pKa = 10.38DD704 pKa = 4.59EE705 pKa = 5.75NGDD708 pKa = 3.75DD709 pKa = 3.38PGYY712 pKa = 10.74YY713 pKa = 10.22VLATIEE719 pKa = 4.4GSDD722 pKa = 4.24TVLTDD727 pKa = 2.96QDD729 pKa = 4.19VVVTPMAA736 pKa = 4.53
MM1 pKa = 7.78SDD3 pKa = 3.05AKK5 pKa = 10.79SAIGTVVKK13 pKa = 10.74ISGQVFAEE21 pKa = 4.34ADD23 pKa = 3.64GHH25 pKa = 5.09RR26 pKa = 11.84HH27 pKa = 5.53PLAEE31 pKa = 4.33GSTVRR36 pKa = 11.84EE37 pKa = 4.34DD38 pKa = 3.66EE39 pKa = 4.55TVVTADD45 pKa = 3.26GSKK48 pKa = 10.32VEE50 pKa = 5.59IRR52 pKa = 11.84FTDD55 pKa = 3.49NTVLSQGEE63 pKa = 3.95NSRR66 pKa = 11.84IEE68 pKa = 3.92LDD70 pKa = 3.44EE71 pKa = 3.89YY72 pKa = 11.43VYY74 pKa = 11.1EE75 pKa = 4.44GDD77 pKa = 3.83KK78 pKa = 11.18GAAGLLFNMVQGSFRR93 pKa = 11.84TVTGKK98 pKa = 9.85IVEE101 pKa = 4.18QNPEE105 pKa = 4.07GFNLSSPLATIGIRR119 pKa = 11.84GTTTFHH125 pKa = 6.99QIGPDD130 pKa = 3.85GEE132 pKa = 4.22RR133 pKa = 11.84HH134 pKa = 4.88GVQNIANPSLGGLDD148 pKa = 3.82SPLHH152 pKa = 5.57GMPSGYY158 pKa = 8.48HH159 pKa = 4.76TMLVTFSNGEE169 pKa = 3.89TRR171 pKa = 11.84VIADD175 pKa = 4.2SYY177 pKa = 11.67QMVDD181 pKa = 3.46LTSAGIGMIRR191 pKa = 11.84TFSLNEE197 pKa = 3.78LLDD200 pKa = 4.87FIDD203 pKa = 5.33LSAHH207 pKa = 5.28SPEE210 pKa = 4.15EE211 pKa = 4.09RR212 pKa = 11.84EE213 pKa = 4.32SLVDD217 pKa = 3.51TLLHH221 pKa = 6.16EE222 pKa = 5.0FRR224 pKa = 11.84EE225 pKa = 4.39QVPPEE230 pKa = 4.25PQSEE234 pKa = 4.42HH235 pKa = 5.94QSSSGDD241 pKa = 3.44NLSPDD246 pKa = 3.35VTDD249 pKa = 6.37DD250 pKa = 3.47GTQDD254 pKa = 3.91SDD256 pKa = 5.74GEE258 pKa = 4.14DD259 pKa = 3.09QQAQDD264 pKa = 4.45EE265 pKa = 4.8VPPDD269 pKa = 3.39IQRR272 pKa = 11.84IISDD276 pKa = 4.32PLPEE280 pKa = 4.14PVIQGGGGNGGGWNPPQEE298 pKa = 4.77DD299 pKa = 4.11PPVRR303 pKa = 11.84DD304 pKa = 3.67VIEE307 pKa = 4.63IIPRR311 pKa = 11.84ITFPIFLIGDD321 pKa = 4.28GGNNSIEE328 pKa = 4.06GTEE331 pKa = 4.13YY332 pKa = 10.39PDD334 pKa = 5.61FIQGLAGDD342 pKa = 4.18DD343 pKa = 4.09SLWGLGSNDD352 pKa = 3.38TLQGGEE358 pKa = 4.49GNDD361 pKa = 3.53VLYY364 pKa = 11.01GGAGDD369 pKa = 4.75DD370 pKa = 3.77VLDD373 pKa = 4.5GGRR376 pKa = 11.84GDD378 pKa = 3.68NSIDD382 pKa = 3.6GGAGNNWVSYY392 pKa = 10.98ASVRR396 pKa = 11.84DD397 pKa = 3.6TFQNKK402 pKa = 8.8GVVVYY407 pKa = 10.81LDD409 pKa = 4.61DD410 pKa = 5.39EE411 pKa = 6.25GDD413 pKa = 3.31DD414 pKa = 3.89SYY416 pKa = 12.02GVRR419 pKa = 11.84EE420 pKa = 4.08YY421 pKa = 10.35PVYY424 pKa = 10.44EE425 pKa = 4.29SNGNLLSEE433 pKa = 4.29PHH435 pKa = 6.3SEE437 pKa = 4.05YY438 pKa = 11.2DD439 pKa = 3.91DD440 pKa = 4.2LASIANVEE448 pKa = 4.05GSIFSDD454 pKa = 4.13AIYY457 pKa = 10.83GNDD460 pKa = 4.01HH461 pKa = 6.43NNHH464 pKa = 6.17LKK466 pKa = 10.72GLAGDD471 pKa = 4.39DD472 pKa = 4.32SIIGNAGDD480 pKa = 3.73DD481 pKa = 4.12TLDD484 pKa = 3.79GGQGNDD490 pKa = 4.06TIDD493 pKa = 3.64GGLGSDD499 pKa = 4.12VIIGGSGNDD508 pKa = 3.06WVSYY512 pKa = 9.35QSYY515 pKa = 9.69YY516 pKa = 10.63SCSFQGMSIDD526 pKa = 4.38LVNGACSLDD535 pKa = 3.63TSSEE539 pKa = 4.06DD540 pKa = 3.25TDD542 pKa = 3.45QFSGIEE548 pKa = 3.95NVEE551 pKa = 3.72GSEE554 pKa = 4.27YY555 pKa = 10.92GDD557 pKa = 4.36EE558 pKa = 4.19ICGDD562 pKa = 3.65SLGNTLHH569 pKa = 6.79GMGGSDD575 pKa = 4.27TISGGAGSDD584 pKa = 3.95FINGGEE590 pKa = 4.97GIDD593 pKa = 3.6TLYY596 pKa = 11.46GNAGADD602 pKa = 3.08TFYY605 pKa = 10.75FDD607 pKa = 4.37NMEE610 pKa = 5.12DD611 pKa = 3.63GFSVDD616 pKa = 2.93THH618 pKa = 8.34ISFSGQQGDD627 pKa = 4.3LIKK630 pKa = 10.95DD631 pKa = 3.79FTPGQDD637 pKa = 4.05KK638 pKa = 10.81ILLDD642 pKa = 3.71SSQYY646 pKa = 9.65SQLGMVVSYY655 pKa = 7.96DD656 pKa = 3.62TNFITIDD663 pKa = 3.37GLYY666 pKa = 10.45DD667 pKa = 3.47GNNATDD673 pKa = 3.37VQGAYY678 pKa = 10.46SSWLNGEE685 pKa = 3.68ACLIVDD691 pKa = 3.76RR692 pKa = 11.84DD693 pKa = 4.01YY694 pKa = 12.18VNDD697 pKa = 4.04CCRR700 pKa = 11.84LIYY703 pKa = 10.38DD704 pKa = 4.59EE705 pKa = 5.75NGDD708 pKa = 3.75DD709 pKa = 3.38PGYY712 pKa = 10.74YY713 pKa = 10.22VLATIEE719 pKa = 4.4GSDD722 pKa = 4.24TVLTDD727 pKa = 2.96QDD729 pKa = 4.19VVVTPMAA736 pKa = 4.53
Molecular weight: 78.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F3YZS7|F3YZS7_DESAF Putative PAS/PAC sensor protein OS=Desulfocurvibacter africanus subsp. africanus str. Walvis Bay OX=690850 GN=Desaf_2561 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.09QPSKK10 pKa = 9.44IKK12 pKa = 10.44RR13 pKa = 11.84KK14 pKa = 7.56RR15 pKa = 11.84THH17 pKa = 5.78GFRR20 pKa = 11.84VRR22 pKa = 11.84MSTKK26 pKa = 10.1NGRR29 pKa = 11.84NMLNRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.05GRR40 pKa = 11.84QRR42 pKa = 11.84LSAA45 pKa = 4.04
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.09QPSKK10 pKa = 9.44IKK12 pKa = 10.44RR13 pKa = 11.84KK14 pKa = 7.56RR15 pKa = 11.84THH17 pKa = 5.78GFRR20 pKa = 11.84VRR22 pKa = 11.84MSTKK26 pKa = 10.1NGRR29 pKa = 11.84NMLNRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.05GRR40 pKa = 11.84QRR42 pKa = 11.84LSAA45 pKa = 4.04
Molecular weight: 5.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1178996 |
30 |
1935 |
318.7 |
35.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.552 ± 0.05 | 1.253 ± 0.018 |
5.273 ± 0.026 | 6.573 ± 0.035 |
3.808 ± 0.025 | 8.201 ± 0.032 |
2.037 ± 0.019 | 4.858 ± 0.033 |
4.044 ± 0.038 | 11.278 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.713 ± 0.021 | 2.633 ± 0.019 |
4.897 ± 0.029 | 3.536 ± 0.027 |
7.019 ± 0.034 | 5.667 ± 0.024 |
4.792 ± 0.026 | 7.147 ± 0.029 |
1.245 ± 0.017 | 2.474 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
