
Modestobacter multiseptatus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Modestobacter
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
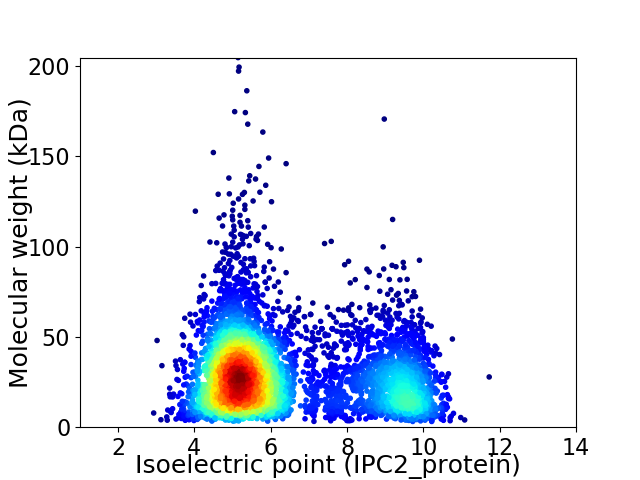
Virtual 2D-PAGE plot for 5324 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A543CAX5|A0A543CAX5_9ACTN Nicotinamide-nucleotide amidase OS=Modestobacter multiseptatus OX=88139 GN=FB553_3403 PE=4 SV=1
MM1 pKa = 7.3VPATTEE7 pKa = 4.11RR8 pKa = 11.84EE9 pKa = 4.16PMPATSRR16 pKa = 11.84RR17 pKa = 11.84PARR20 pKa = 11.84ALLGGVVAAVVLTGCAPTLVVGSATPAAGQVGEE53 pKa = 4.4VRR55 pKa = 11.84AADD58 pKa = 3.95FPIIGATDD66 pKa = 3.48EE67 pKa = 5.06VVDD70 pKa = 3.92QLARR74 pKa = 11.84NALSDD79 pKa = 4.79LNTYY83 pKa = 7.43WQQQLPDD90 pKa = 3.63TLGTDD95 pKa = 4.22FTGLAGGYY103 pKa = 10.0FSVDD107 pKa = 3.39PANVDD112 pKa = 3.27PAQYY116 pKa = 9.79PGGEE120 pKa = 4.25IGCGEE125 pKa = 4.48PPDD128 pKa = 4.7AVADD132 pKa = 3.73NAFYY136 pKa = 10.75CPPSSEE142 pKa = 4.37YY143 pKa = 10.44PNGDD147 pKa = 3.79AIQYY151 pKa = 9.28DD152 pKa = 3.86RR153 pKa = 11.84SFLRR157 pKa = 11.84EE158 pKa = 3.62LADD161 pKa = 3.99DD162 pKa = 3.86YY163 pKa = 11.9GRR165 pKa = 11.84FLPALVMAHH174 pKa = 6.23EE175 pKa = 5.31FGHH178 pKa = 6.69AVQARR183 pKa = 11.84VGYY186 pKa = 8.75PLQASIAIEE195 pKa = 4.03TQADD199 pKa = 3.98CFAGAWTAWVADD211 pKa = 3.86GNAPHH216 pKa = 5.84TTIRR220 pKa = 11.84TPEE223 pKa = 4.18LDD225 pKa = 3.36EE226 pKa = 4.06VLRR229 pKa = 11.84GYY231 pKa = 10.87SLLRR235 pKa = 11.84DD236 pKa = 3.59PVGTSVNEE244 pKa = 4.28TEE246 pKa = 4.21AHH248 pKa = 6.12GSLFDD253 pKa = 4.09RR254 pKa = 11.84VSAFQDD260 pKa = 3.6GFDD263 pKa = 5.04DD264 pKa = 5.52GPGACRR270 pKa = 11.84DD271 pKa = 3.73DD272 pKa = 4.58FGADD276 pKa = 3.01RR277 pKa = 11.84PYY279 pKa = 10.1TQSEE283 pKa = 4.5FNPAEE288 pKa = 3.95AANNGDD294 pKa = 2.79ASYY297 pKa = 10.58PVTQDD302 pKa = 4.66LITTALPDD310 pKa = 3.9FWTSAFSGLDD320 pKa = 3.29EE321 pKa = 4.25QFQAPVIVPFSGTAPDD337 pKa = 3.68CGGRR341 pKa = 11.84EE342 pKa = 4.16PDD344 pKa = 3.39TDD346 pKa = 4.59LVFCAADD353 pKa = 3.41DD354 pKa = 3.96TVGYY358 pKa = 10.78DD359 pKa = 3.73EE360 pKa = 5.47TDD362 pKa = 3.22LAQPVYY368 pKa = 11.21ADD370 pKa = 3.53LGDD373 pKa = 3.99YY374 pKa = 10.98AVLTAAAIPYY384 pKa = 9.85SLAARR389 pKa = 11.84DD390 pKa = 3.82QLGLSTDD397 pKa = 3.58DD398 pKa = 3.49QDD400 pKa = 4.2ALRR403 pKa = 11.84SAICLTGAFTFAVLRR418 pKa = 11.84GSVSAIQISPGDD430 pKa = 3.41VDD432 pKa = 4.16EE433 pKa = 5.08SVQFLLEE440 pKa = 4.07YY441 pKa = 10.8SDD443 pKa = 5.35DD444 pKa = 3.94PTVLADD450 pKa = 3.68GGLTGFQLVDD460 pKa = 3.73LFRR463 pKa = 11.84SGVFGGLSSCGIGGG477 pKa = 3.51
MM1 pKa = 7.3VPATTEE7 pKa = 4.11RR8 pKa = 11.84EE9 pKa = 4.16PMPATSRR16 pKa = 11.84RR17 pKa = 11.84PARR20 pKa = 11.84ALLGGVVAAVVLTGCAPTLVVGSATPAAGQVGEE53 pKa = 4.4VRR55 pKa = 11.84AADD58 pKa = 3.95FPIIGATDD66 pKa = 3.48EE67 pKa = 5.06VVDD70 pKa = 3.92QLARR74 pKa = 11.84NALSDD79 pKa = 4.79LNTYY83 pKa = 7.43WQQQLPDD90 pKa = 3.63TLGTDD95 pKa = 4.22FTGLAGGYY103 pKa = 10.0FSVDD107 pKa = 3.39PANVDD112 pKa = 3.27PAQYY116 pKa = 9.79PGGEE120 pKa = 4.25IGCGEE125 pKa = 4.48PPDD128 pKa = 4.7AVADD132 pKa = 3.73NAFYY136 pKa = 10.75CPPSSEE142 pKa = 4.37YY143 pKa = 10.44PNGDD147 pKa = 3.79AIQYY151 pKa = 9.28DD152 pKa = 3.86RR153 pKa = 11.84SFLRR157 pKa = 11.84EE158 pKa = 3.62LADD161 pKa = 3.99DD162 pKa = 3.86YY163 pKa = 11.9GRR165 pKa = 11.84FLPALVMAHH174 pKa = 6.23EE175 pKa = 5.31FGHH178 pKa = 6.69AVQARR183 pKa = 11.84VGYY186 pKa = 8.75PLQASIAIEE195 pKa = 4.03TQADD199 pKa = 3.98CFAGAWTAWVADD211 pKa = 3.86GNAPHH216 pKa = 5.84TTIRR220 pKa = 11.84TPEE223 pKa = 4.18LDD225 pKa = 3.36EE226 pKa = 4.06VLRR229 pKa = 11.84GYY231 pKa = 10.87SLLRR235 pKa = 11.84DD236 pKa = 3.59PVGTSVNEE244 pKa = 4.28TEE246 pKa = 4.21AHH248 pKa = 6.12GSLFDD253 pKa = 4.09RR254 pKa = 11.84VSAFQDD260 pKa = 3.6GFDD263 pKa = 5.04DD264 pKa = 5.52GPGACRR270 pKa = 11.84DD271 pKa = 3.73DD272 pKa = 4.58FGADD276 pKa = 3.01RR277 pKa = 11.84PYY279 pKa = 10.1TQSEE283 pKa = 4.5FNPAEE288 pKa = 3.95AANNGDD294 pKa = 2.79ASYY297 pKa = 10.58PVTQDD302 pKa = 4.66LITTALPDD310 pKa = 3.9FWTSAFSGLDD320 pKa = 3.29EE321 pKa = 4.25QFQAPVIVPFSGTAPDD337 pKa = 3.68CGGRR341 pKa = 11.84EE342 pKa = 4.16PDD344 pKa = 3.39TDD346 pKa = 4.59LVFCAADD353 pKa = 3.41DD354 pKa = 3.96TVGYY358 pKa = 10.78DD359 pKa = 3.73EE360 pKa = 5.47TDD362 pKa = 3.22LAQPVYY368 pKa = 11.21ADD370 pKa = 3.53LGDD373 pKa = 3.99YY374 pKa = 10.98AVLTAAAIPYY384 pKa = 9.85SLAARR389 pKa = 11.84DD390 pKa = 3.82QLGLSTDD397 pKa = 3.58DD398 pKa = 3.49QDD400 pKa = 4.2ALRR403 pKa = 11.84SAICLTGAFTFAVLRR418 pKa = 11.84GSVSAIQISPGDD430 pKa = 3.41VDD432 pKa = 4.16EE433 pKa = 5.08SVQFLLEE440 pKa = 4.07YY441 pKa = 10.8SDD443 pKa = 5.35DD444 pKa = 3.94PTVLADD450 pKa = 3.68GGLTGFQLVDD460 pKa = 3.73LFRR463 pKa = 11.84SGVFGGLSSCGIGGG477 pKa = 3.51
Molecular weight: 50.1 kDa
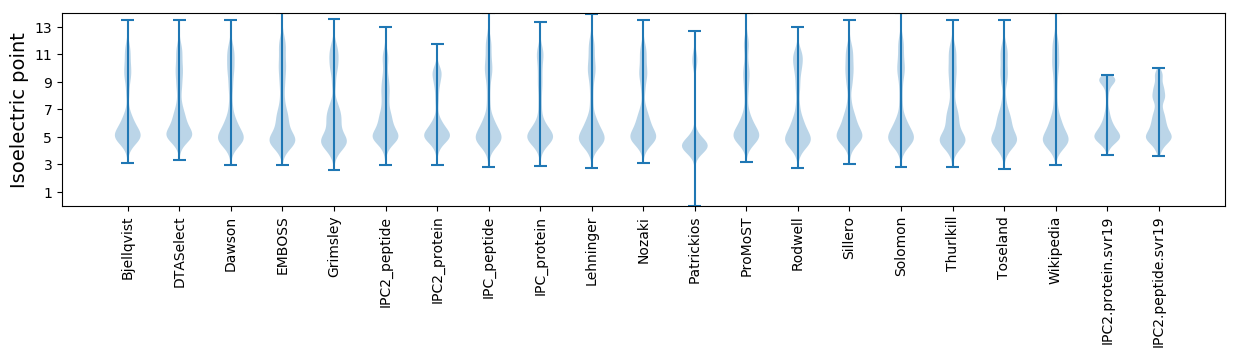
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A543C4M6|A0A543C4M6_9ACTN Uncharacterized protein OS=Modestobacter multiseptatus OX=88139 GN=FB553_0967 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1567677 |
29 |
1939 |
294.5 |
31.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.263 ± 0.053 | 0.747 ± 0.011 |
6.128 ± 0.028 | 5.099 ± 0.033 |
2.517 ± 0.019 | 9.559 ± 0.032 |
2.085 ± 0.015 | 3.004 ± 0.024 |
1.479 ± 0.021 | 10.617 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.657 ± 0.012 | 1.56 ± 0.017 |
6.161 ± 0.03 | 2.965 ± 0.021 |
8.147 ± 0.034 | 5.097 ± 0.024 |
6.183 ± 0.03 | 9.45 ± 0.033 |
1.498 ± 0.016 | 1.783 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
