
Lactobacillus collinoides DSM 20515 = JCM 1123
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Secundilactobacillus; Secundilactobacillus collinoides
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
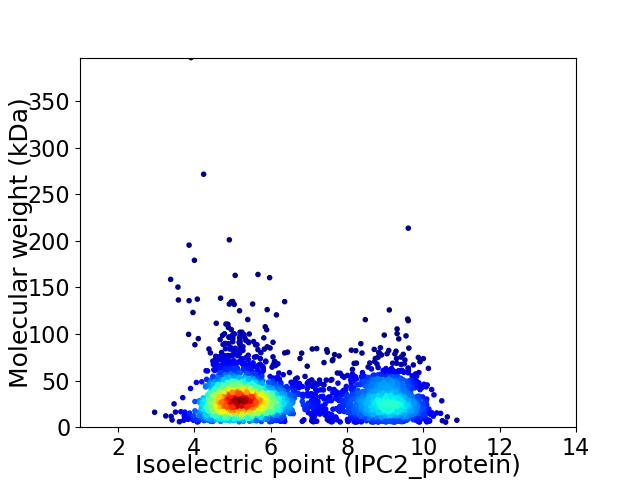
Virtual 2D-PAGE plot for 3157 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R2B9U1|A0A0R2B9U1_LACCL Branched-chain amino acid ABC transporter substrate binding protein OS=Lactobacillus collinoides DSM 20515 = JCM 1123 OX=1423733 GN=FC82_GL002087 PE=3 SV=1
MM1 pKa = 8.04DD2 pKa = 5.11AGKK5 pKa = 8.0TVPEE9 pKa = 4.31TTKK12 pKa = 10.31TPTGGISSASSNTDD26 pKa = 2.43SSSILDD32 pKa = 3.71EE33 pKa = 4.78NKK35 pKa = 9.65TEE37 pKa = 4.17TPSVIDD43 pKa = 4.09DD44 pKa = 3.82VPGEE48 pKa = 4.14VSGTPTIADD57 pKa = 3.23DD58 pKa = 3.79TLNVNKK64 pKa = 9.0LTLINPTEE72 pKa = 4.17KK73 pKa = 10.75EE74 pKa = 3.03IDD76 pKa = 3.42AAKK79 pKa = 9.22EE80 pKa = 4.09TMEE83 pKa = 4.56AAYY86 pKa = 9.69KK87 pKa = 9.55LTGQAQQLNALLDD100 pKa = 3.69EE101 pKa = 5.0AAPSASSAVITLTSNTDD118 pKa = 3.32VIGYY122 pKa = 10.0NSGQTSIILTFTVVGAKK139 pKa = 10.4AGDD142 pKa = 3.89VYY144 pKa = 10.91TITIPKK150 pKa = 8.98TSVFSYY156 pKa = 11.3SSMDD160 pKa = 3.58KK161 pKa = 10.21LTATQGTSEE170 pKa = 4.32TQSTADD176 pKa = 3.78GGTMITVTMARR187 pKa = 11.84SVSDD191 pKa = 3.44LSFKK195 pKa = 10.65IKK197 pKa = 10.64LGTANNEE204 pKa = 4.07NQQPRR209 pKa = 11.84PMSDD213 pKa = 2.54VGVNTYY219 pKa = 9.88TITAMNGDD227 pKa = 4.12TEE229 pKa = 4.49VASIPITQTITPTISMGNVTRR250 pKa = 11.84TTPNLSEE257 pKa = 4.06LTAVQSGVDD266 pKa = 3.33YY267 pKa = 10.85IYY269 pKa = 10.33TLKK272 pKa = 10.95VNEE275 pKa = 4.28ADD277 pKa = 3.35GVYY280 pKa = 10.7NDD282 pKa = 5.0GSTSSYY288 pKa = 10.85VNAAVNYY295 pKa = 7.06GTTITIPVPEE305 pKa = 4.72GFTLNSDD312 pKa = 3.61VTNSINGFSDD322 pKa = 3.27EE323 pKa = 4.45TTISQPDD330 pKa = 3.7GSGTNVIITVPKK342 pKa = 9.94GSGKK346 pKa = 7.92EE347 pKa = 4.11TYY349 pKa = 9.95EE350 pKa = 4.1GGSGYY355 pKa = 10.89KK356 pKa = 10.26LVGSYY361 pKa = 10.71DD362 pKa = 3.5VAASEE367 pKa = 5.02SEE369 pKa = 4.14QTLTASGPVTITQVIDD385 pKa = 3.54GTGTTITDD393 pKa = 3.13EE394 pKa = 4.4SEE396 pKa = 4.22NVFSEE401 pKa = 4.19TLAASTEE408 pKa = 4.06IPDD411 pKa = 3.95ASNISISMHH420 pKa = 5.94GNSSSTSTEE429 pKa = 3.6LALDD433 pKa = 3.99GDD435 pKa = 4.34TTNDD439 pKa = 3.21PKK441 pKa = 11.52YY442 pKa = 9.57LTTYY446 pKa = 11.11SLTDD450 pKa = 3.24NSAISLTDD458 pKa = 3.86DD459 pKa = 3.89LNLTINVPDD468 pKa = 4.96GIEE471 pKa = 4.07MTSLTVPAEE480 pKa = 4.2GATTSQYY487 pKa = 11.44LPGTTSYY494 pKa = 11.31AYY496 pKa = 10.49QLTLADD502 pKa = 4.11GSIEE506 pKa = 3.98NGEE509 pKa = 3.95VDD511 pKa = 3.78AGGTITANYY520 pKa = 8.66GSEE523 pKa = 3.82VRR525 pKa = 11.84QIVLTPNYY533 pKa = 9.16LAAGAYY539 pKa = 7.02TSSNAFILSGIMSSKK554 pKa = 10.71YY555 pKa = 10.46DD556 pKa = 3.87DD557 pKa = 4.73GSDD560 pKa = 3.39VQYY563 pKa = 11.49GDD565 pKa = 4.41TIDD568 pKa = 4.01FTSSLALSSDD578 pKa = 3.17PSTIISSDD586 pKa = 3.75SIKK589 pKa = 9.61QTVIEE594 pKa = 4.53SFAHH598 pKa = 6.24FYY600 pKa = 10.94AYY602 pKa = 10.78LSQSDD607 pKa = 3.93KK608 pKa = 11.31TPGAVAAGSLSLRR621 pKa = 11.84MTGQDD626 pKa = 2.41SHH628 pKa = 5.45TTYY631 pKa = 10.74EE632 pKa = 3.97VYY634 pKa = 10.58EE635 pKa = 4.4PIFYY639 pKa = 10.66YY640 pKa = 10.46VLPSATYY647 pKa = 9.33ISGFSAIDD655 pKa = 3.31SKK657 pKa = 11.97AKK659 pKa = 9.03VTEE662 pKa = 4.39FQADD666 pKa = 3.53DD667 pKa = 3.64GRR669 pKa = 11.84TVVQIDD675 pKa = 3.61YY676 pKa = 10.35SGTGEE681 pKa = 4.12YY682 pKa = 10.92VSTNSTFSQQVKK694 pKa = 9.75LSNYY698 pKa = 9.97ADD700 pKa = 4.48ALPGSYY706 pKa = 10.4GYY708 pKa = 11.46LMYY711 pKa = 10.56VYY713 pKa = 10.88SPVTKK718 pKa = 10.45LANTKK723 pKa = 9.56MISDD727 pKa = 4.27TSYY730 pKa = 10.73SAGNAEE736 pKa = 3.85AVEE739 pKa = 4.56FGSGNWTISSASTFDD754 pKa = 3.29QYY756 pKa = 11.99SFGQTNEE763 pKa = 3.96DD764 pKa = 3.5VQGVTNATSDD774 pKa = 3.78DD775 pKa = 3.95QGSSDD780 pKa = 3.15ATFYY784 pKa = 10.96VALVNTLRR792 pKa = 11.84DD793 pKa = 3.72GTDD796 pKa = 2.9NTTTSIINLPTSDD809 pKa = 4.24DD810 pKa = 4.11ANGSGYY816 pKa = 8.74TFNLTGPVTLPTNFITATGTGDD838 pKa = 3.69TIDD841 pKa = 3.54ATVLYY846 pKa = 9.04STATVAVTQGQKK858 pKa = 9.74TIDD861 pKa = 3.26TSSYY865 pKa = 9.44VSADD869 pKa = 3.35EE870 pKa = 4.14VTSWDD875 pKa = 3.79SIRR878 pKa = 11.84SVLIKK883 pKa = 10.38INNFGLDD890 pKa = 3.48DD891 pKa = 3.99STGKK895 pKa = 10.04IALNGTIADD904 pKa = 4.02MQNLAGMTGYY914 pKa = 10.23LQAIVLVNDD923 pKa = 4.67DD924 pKa = 4.18GASVDD929 pKa = 3.54NSTSLSITGTSTVTARR945 pKa = 11.84FHH947 pKa = 6.9YY948 pKa = 10.31IDD950 pKa = 5.76DD951 pKa = 4.18NGEE954 pKa = 3.93DD955 pKa = 3.63QYY957 pKa = 11.12ITLDD961 pKa = 3.84DD962 pKa = 4.72LKK964 pKa = 10.55QTDD967 pKa = 3.82LQDD970 pKa = 4.12NVDD973 pKa = 4.35SLQDD977 pKa = 3.42IYY979 pKa = 10.35PTNLDD984 pKa = 3.27GFSAVDD990 pKa = 3.2QALIPEE996 pKa = 5.57GYY998 pKa = 8.25TLASNQYY1005 pKa = 8.93TYY1007 pKa = 11.68VSGANADD1014 pKa = 4.51GIMSSATTKK1023 pKa = 10.81VGDD1026 pKa = 3.6TAVYY1030 pKa = 8.51NTDD1033 pKa = 3.4GEE1035 pKa = 4.49IVQYY1039 pKa = 10.61EE1040 pKa = 4.15LAKK1043 pKa = 10.6NASLTVTYY1051 pKa = 10.23IDD1053 pKa = 4.56DD1054 pKa = 4.26AADD1057 pKa = 3.76GAVVGDD1063 pKa = 3.74VTTVTGLLGASDD1075 pKa = 4.13TYY1077 pKa = 11.28KK1078 pKa = 10.72VVIPNGYY1085 pKa = 9.35EE1086 pKa = 3.86LASGQASSIGYY1097 pKa = 7.03TLSDD1101 pKa = 3.62SDD1103 pKa = 3.85TDD1105 pKa = 3.43NLTVHH1110 pKa = 7.03LVHH1113 pKa = 6.83QITVIGTKK1121 pKa = 7.55TTTNTVTYY1129 pKa = 9.7TGLPTDD1135 pKa = 4.34KK1136 pKa = 10.69TPASQSKK1143 pKa = 8.84KK1144 pKa = 9.29VSWNVEE1150 pKa = 3.69KK1151 pKa = 10.95DD1152 pKa = 3.56DD1153 pKa = 3.67VTGEE1157 pKa = 4.04VTYY1160 pKa = 10.86VPSSTHH1166 pKa = 3.5QTTYY1170 pKa = 7.75TTTNVSGYY1178 pKa = 10.71YY1179 pKa = 9.55YY1180 pKa = 10.47DD1181 pKa = 4.33SSQAQVGFTVTTQTTEE1197 pKa = 4.02PKK1199 pKa = 10.05DD1200 pKa = 3.48QSIEE1204 pKa = 3.74VAYY1207 pKa = 10.53YY1208 pKa = 11.16AEE1210 pKa = 4.2TQTTNIEE1217 pKa = 4.65FVDD1220 pKa = 4.28DD1221 pKa = 4.55DD1222 pKa = 4.31NNSAVVGTQTVLQGKK1237 pKa = 7.04TDD1239 pKa = 3.79EE1240 pKa = 4.21NVPWSVSVPDD1250 pKa = 4.03NYY1252 pKa = 10.85VLASGQSSSGTYY1264 pKa = 7.57TFKK1267 pKa = 11.15ASDD1270 pKa = 3.53NDD1272 pKa = 3.78NIIVHH1277 pKa = 6.81LSHH1280 pKa = 6.82IQATGTTTTTRR1291 pKa = 11.84NIYY1294 pKa = 10.46FIVEE1298 pKa = 4.33YY1299 pKa = 10.72NNSLNPEE1306 pKa = 4.26PVNQTAMWNVTKK1318 pKa = 11.05DD1319 pKa = 3.46MVTGAITATTTDD1331 pKa = 3.44SYY1333 pKa = 11.66NQVDD1337 pKa = 4.14SLPISGYY1344 pKa = 6.55TANTAIVPSEE1354 pKa = 3.96NLEE1357 pKa = 4.65PITTLPANAADD1368 pKa = 3.55IYY1370 pKa = 9.95VTYY1373 pKa = 9.3TADD1376 pKa = 3.28AQTASTEE1383 pKa = 4.13YY1384 pKa = 10.96VDD1386 pKa = 5.94DD1387 pKa = 4.34SASEE1391 pKa = 4.49TIVGTPKK1398 pKa = 10.36IIDD1401 pKa = 4.43GYY1403 pKa = 9.78TGTSTNWDD1411 pKa = 3.7TSDD1414 pKa = 4.71LPAGYY1419 pKa = 9.72QLAANQPSSGTYY1431 pKa = 9.59TFTAEE1436 pKa = 4.24DD1437 pKa = 3.6NQVVQIHH1444 pKa = 5.48VVHH1447 pKa = 6.58IHH1449 pKa = 5.38TQSEE1453 pKa = 4.78MSTKK1457 pKa = 10.06DD1458 pKa = 2.99TVNYY1462 pKa = 10.6YY1463 pKa = 9.89EE1464 pKa = 4.84LPQDD1468 pKa = 3.64KK1469 pKa = 10.94AEE1471 pKa = 4.0QSKK1474 pKa = 8.83TEE1476 pKa = 4.09SVNWKK1481 pKa = 9.65IDD1483 pKa = 3.3TDD1485 pKa = 3.79EE1486 pKa = 4.25ATGISTFTPSSTAIEE1501 pKa = 4.1ISTPEE1506 pKa = 3.91VPGYY1510 pKa = 8.97TPSEE1514 pKa = 3.9QTIKK1518 pKa = 9.75FTQATSEE1525 pKa = 4.33TQPSDD1530 pKa = 3.0QTADD1534 pKa = 3.33VTYY1537 pKa = 10.65SADD1540 pKa = 3.46PQEE1543 pKa = 4.09ITFTYY1548 pKa = 10.82VDD1550 pKa = 4.25DD1551 pKa = 4.05TTGDD1555 pKa = 3.26EE1556 pKa = 4.53MEE1558 pKa = 4.84NLDD1561 pKa = 3.8ITLEE1565 pKa = 4.09GHH1567 pKa = 5.14TDD1569 pKa = 3.12EE1570 pKa = 5.76AGTYY1574 pKa = 7.63TVIVPKK1580 pKa = 10.63NYY1582 pKa = 9.97VLSQGQSATVPYY1594 pKa = 9.55TYY1596 pKa = 9.9TADD1599 pKa = 3.31VQTPTKK1605 pKa = 10.61VHH1607 pKa = 6.03LTHH1610 pKa = 7.34AIADD1614 pKa = 4.07GTVVTTRR1621 pKa = 11.84TITYY1625 pKa = 8.78VVNGNPTAAPKK1636 pKa = 10.67AVVQKK1641 pKa = 9.32ITWKK1645 pKa = 10.1VVRR1648 pKa = 11.84DD1649 pKa = 3.84MVTGTSYY1656 pKa = 10.23ATAQNGYY1663 pKa = 9.76DD1664 pKa = 3.46AVITPEE1670 pKa = 4.04LAGYY1674 pKa = 7.2TADD1677 pKa = 3.78KK1678 pKa = 11.04SGIAQEE1684 pKa = 3.93LLGRR1688 pKa = 11.84VDD1690 pKa = 3.6TADD1693 pKa = 3.96LADD1696 pKa = 3.72SAVTVTYY1703 pKa = 8.9TAIPVAPDD1711 pKa = 3.15NDD1713 pKa = 4.11SEE1715 pKa = 4.85NEE1717 pKa = 4.37VVTPTQPDD1725 pKa = 3.72TAVSGTMGSDD1735 pKa = 3.09VDD1737 pKa = 4.57NEE1739 pKa = 4.17QSDD1742 pKa = 4.19VTVLEE1747 pKa = 4.36KK1748 pKa = 11.02DD1749 pKa = 3.5NQKK1752 pKa = 8.37VTRR1755 pKa = 11.84DD1756 pKa = 3.64GNSDD1760 pKa = 3.42EE1761 pKa = 4.54TTKK1764 pKa = 11.22SNAFGSSAGIYY1775 pKa = 8.58VHH1777 pKa = 7.06GSTHH1781 pKa = 6.75ADD1783 pKa = 3.01AVPTVTTDD1791 pKa = 3.15DD1792 pKa = 3.71TGVKK1796 pKa = 10.02KK1797 pKa = 9.71GTANKK1802 pKa = 10.11VNITTAAKK1810 pKa = 10.33LPQTDD1815 pKa = 3.68EE1816 pKa = 4.22KK1817 pKa = 11.46QNGIWTIAGASLLGLFSLFGIGKK1840 pKa = 9.22KK1841 pKa = 9.5KK1842 pKa = 10.27RR1843 pKa = 11.84RR1844 pKa = 11.84EE1845 pKa = 3.71DD1846 pKa = 3.51DD1847 pKa = 3.16
MM1 pKa = 8.04DD2 pKa = 5.11AGKK5 pKa = 8.0TVPEE9 pKa = 4.31TTKK12 pKa = 10.31TPTGGISSASSNTDD26 pKa = 2.43SSSILDD32 pKa = 3.71EE33 pKa = 4.78NKK35 pKa = 9.65TEE37 pKa = 4.17TPSVIDD43 pKa = 4.09DD44 pKa = 3.82VPGEE48 pKa = 4.14VSGTPTIADD57 pKa = 3.23DD58 pKa = 3.79TLNVNKK64 pKa = 9.0LTLINPTEE72 pKa = 4.17KK73 pKa = 10.75EE74 pKa = 3.03IDD76 pKa = 3.42AAKK79 pKa = 9.22EE80 pKa = 4.09TMEE83 pKa = 4.56AAYY86 pKa = 9.69KK87 pKa = 9.55LTGQAQQLNALLDD100 pKa = 3.69EE101 pKa = 5.0AAPSASSAVITLTSNTDD118 pKa = 3.32VIGYY122 pKa = 10.0NSGQTSIILTFTVVGAKK139 pKa = 10.4AGDD142 pKa = 3.89VYY144 pKa = 10.91TITIPKK150 pKa = 8.98TSVFSYY156 pKa = 11.3SSMDD160 pKa = 3.58KK161 pKa = 10.21LTATQGTSEE170 pKa = 4.32TQSTADD176 pKa = 3.78GGTMITVTMARR187 pKa = 11.84SVSDD191 pKa = 3.44LSFKK195 pKa = 10.65IKK197 pKa = 10.64LGTANNEE204 pKa = 4.07NQQPRR209 pKa = 11.84PMSDD213 pKa = 2.54VGVNTYY219 pKa = 9.88TITAMNGDD227 pKa = 4.12TEE229 pKa = 4.49VASIPITQTITPTISMGNVTRR250 pKa = 11.84TTPNLSEE257 pKa = 4.06LTAVQSGVDD266 pKa = 3.33YY267 pKa = 10.85IYY269 pKa = 10.33TLKK272 pKa = 10.95VNEE275 pKa = 4.28ADD277 pKa = 3.35GVYY280 pKa = 10.7NDD282 pKa = 5.0GSTSSYY288 pKa = 10.85VNAAVNYY295 pKa = 7.06GTTITIPVPEE305 pKa = 4.72GFTLNSDD312 pKa = 3.61VTNSINGFSDD322 pKa = 3.27EE323 pKa = 4.45TTISQPDD330 pKa = 3.7GSGTNVIITVPKK342 pKa = 9.94GSGKK346 pKa = 7.92EE347 pKa = 4.11TYY349 pKa = 9.95EE350 pKa = 4.1GGSGYY355 pKa = 10.89KK356 pKa = 10.26LVGSYY361 pKa = 10.71DD362 pKa = 3.5VAASEE367 pKa = 5.02SEE369 pKa = 4.14QTLTASGPVTITQVIDD385 pKa = 3.54GTGTTITDD393 pKa = 3.13EE394 pKa = 4.4SEE396 pKa = 4.22NVFSEE401 pKa = 4.19TLAASTEE408 pKa = 4.06IPDD411 pKa = 3.95ASNISISMHH420 pKa = 5.94GNSSSTSTEE429 pKa = 3.6LALDD433 pKa = 3.99GDD435 pKa = 4.34TTNDD439 pKa = 3.21PKK441 pKa = 11.52YY442 pKa = 9.57LTTYY446 pKa = 11.11SLTDD450 pKa = 3.24NSAISLTDD458 pKa = 3.86DD459 pKa = 3.89LNLTINVPDD468 pKa = 4.96GIEE471 pKa = 4.07MTSLTVPAEE480 pKa = 4.2GATTSQYY487 pKa = 11.44LPGTTSYY494 pKa = 11.31AYY496 pKa = 10.49QLTLADD502 pKa = 4.11GSIEE506 pKa = 3.98NGEE509 pKa = 3.95VDD511 pKa = 3.78AGGTITANYY520 pKa = 8.66GSEE523 pKa = 3.82VRR525 pKa = 11.84QIVLTPNYY533 pKa = 9.16LAAGAYY539 pKa = 7.02TSSNAFILSGIMSSKK554 pKa = 10.71YY555 pKa = 10.46DD556 pKa = 3.87DD557 pKa = 4.73GSDD560 pKa = 3.39VQYY563 pKa = 11.49GDD565 pKa = 4.41TIDD568 pKa = 4.01FTSSLALSSDD578 pKa = 3.17PSTIISSDD586 pKa = 3.75SIKK589 pKa = 9.61QTVIEE594 pKa = 4.53SFAHH598 pKa = 6.24FYY600 pKa = 10.94AYY602 pKa = 10.78LSQSDD607 pKa = 3.93KK608 pKa = 11.31TPGAVAAGSLSLRR621 pKa = 11.84MTGQDD626 pKa = 2.41SHH628 pKa = 5.45TTYY631 pKa = 10.74EE632 pKa = 3.97VYY634 pKa = 10.58EE635 pKa = 4.4PIFYY639 pKa = 10.66YY640 pKa = 10.46VLPSATYY647 pKa = 9.33ISGFSAIDD655 pKa = 3.31SKK657 pKa = 11.97AKK659 pKa = 9.03VTEE662 pKa = 4.39FQADD666 pKa = 3.53DD667 pKa = 3.64GRR669 pKa = 11.84TVVQIDD675 pKa = 3.61YY676 pKa = 10.35SGTGEE681 pKa = 4.12YY682 pKa = 10.92VSTNSTFSQQVKK694 pKa = 9.75LSNYY698 pKa = 9.97ADD700 pKa = 4.48ALPGSYY706 pKa = 10.4GYY708 pKa = 11.46LMYY711 pKa = 10.56VYY713 pKa = 10.88SPVTKK718 pKa = 10.45LANTKK723 pKa = 9.56MISDD727 pKa = 4.27TSYY730 pKa = 10.73SAGNAEE736 pKa = 3.85AVEE739 pKa = 4.56FGSGNWTISSASTFDD754 pKa = 3.29QYY756 pKa = 11.99SFGQTNEE763 pKa = 3.96DD764 pKa = 3.5VQGVTNATSDD774 pKa = 3.78DD775 pKa = 3.95QGSSDD780 pKa = 3.15ATFYY784 pKa = 10.96VALVNTLRR792 pKa = 11.84DD793 pKa = 3.72GTDD796 pKa = 2.9NTTTSIINLPTSDD809 pKa = 4.24DD810 pKa = 4.11ANGSGYY816 pKa = 8.74TFNLTGPVTLPTNFITATGTGDD838 pKa = 3.69TIDD841 pKa = 3.54ATVLYY846 pKa = 9.04STATVAVTQGQKK858 pKa = 9.74TIDD861 pKa = 3.26TSSYY865 pKa = 9.44VSADD869 pKa = 3.35EE870 pKa = 4.14VTSWDD875 pKa = 3.79SIRR878 pKa = 11.84SVLIKK883 pKa = 10.38INNFGLDD890 pKa = 3.48DD891 pKa = 3.99STGKK895 pKa = 10.04IALNGTIADD904 pKa = 4.02MQNLAGMTGYY914 pKa = 10.23LQAIVLVNDD923 pKa = 4.67DD924 pKa = 4.18GASVDD929 pKa = 3.54NSTSLSITGTSTVTARR945 pKa = 11.84FHH947 pKa = 6.9YY948 pKa = 10.31IDD950 pKa = 5.76DD951 pKa = 4.18NGEE954 pKa = 3.93DD955 pKa = 3.63QYY957 pKa = 11.12ITLDD961 pKa = 3.84DD962 pKa = 4.72LKK964 pKa = 10.55QTDD967 pKa = 3.82LQDD970 pKa = 4.12NVDD973 pKa = 4.35SLQDD977 pKa = 3.42IYY979 pKa = 10.35PTNLDD984 pKa = 3.27GFSAVDD990 pKa = 3.2QALIPEE996 pKa = 5.57GYY998 pKa = 8.25TLASNQYY1005 pKa = 8.93TYY1007 pKa = 11.68VSGANADD1014 pKa = 4.51GIMSSATTKK1023 pKa = 10.81VGDD1026 pKa = 3.6TAVYY1030 pKa = 8.51NTDD1033 pKa = 3.4GEE1035 pKa = 4.49IVQYY1039 pKa = 10.61EE1040 pKa = 4.15LAKK1043 pKa = 10.6NASLTVTYY1051 pKa = 10.23IDD1053 pKa = 4.56DD1054 pKa = 4.26AADD1057 pKa = 3.76GAVVGDD1063 pKa = 3.74VTTVTGLLGASDD1075 pKa = 4.13TYY1077 pKa = 11.28KK1078 pKa = 10.72VVIPNGYY1085 pKa = 9.35EE1086 pKa = 3.86LASGQASSIGYY1097 pKa = 7.03TLSDD1101 pKa = 3.62SDD1103 pKa = 3.85TDD1105 pKa = 3.43NLTVHH1110 pKa = 7.03LVHH1113 pKa = 6.83QITVIGTKK1121 pKa = 7.55TTTNTVTYY1129 pKa = 9.7TGLPTDD1135 pKa = 4.34KK1136 pKa = 10.69TPASQSKK1143 pKa = 8.84KK1144 pKa = 9.29VSWNVEE1150 pKa = 3.69KK1151 pKa = 10.95DD1152 pKa = 3.56DD1153 pKa = 3.67VTGEE1157 pKa = 4.04VTYY1160 pKa = 10.86VPSSTHH1166 pKa = 3.5QTTYY1170 pKa = 7.75TTTNVSGYY1178 pKa = 10.71YY1179 pKa = 9.55YY1180 pKa = 10.47DD1181 pKa = 4.33SSQAQVGFTVTTQTTEE1197 pKa = 4.02PKK1199 pKa = 10.05DD1200 pKa = 3.48QSIEE1204 pKa = 3.74VAYY1207 pKa = 10.53YY1208 pKa = 11.16AEE1210 pKa = 4.2TQTTNIEE1217 pKa = 4.65FVDD1220 pKa = 4.28DD1221 pKa = 4.55DD1222 pKa = 4.31NNSAVVGTQTVLQGKK1237 pKa = 7.04TDD1239 pKa = 3.79EE1240 pKa = 4.21NVPWSVSVPDD1250 pKa = 4.03NYY1252 pKa = 10.85VLASGQSSSGTYY1264 pKa = 7.57TFKK1267 pKa = 11.15ASDD1270 pKa = 3.53NDD1272 pKa = 3.78NIIVHH1277 pKa = 6.81LSHH1280 pKa = 6.82IQATGTTTTTRR1291 pKa = 11.84NIYY1294 pKa = 10.46FIVEE1298 pKa = 4.33YY1299 pKa = 10.72NNSLNPEE1306 pKa = 4.26PVNQTAMWNVTKK1318 pKa = 11.05DD1319 pKa = 3.46MVTGAITATTTDD1331 pKa = 3.44SYY1333 pKa = 11.66NQVDD1337 pKa = 4.14SLPISGYY1344 pKa = 6.55TANTAIVPSEE1354 pKa = 3.96NLEE1357 pKa = 4.65PITTLPANAADD1368 pKa = 3.55IYY1370 pKa = 9.95VTYY1373 pKa = 9.3TADD1376 pKa = 3.28AQTASTEE1383 pKa = 4.13YY1384 pKa = 10.96VDD1386 pKa = 5.94DD1387 pKa = 4.34SASEE1391 pKa = 4.49TIVGTPKK1398 pKa = 10.36IIDD1401 pKa = 4.43GYY1403 pKa = 9.78TGTSTNWDD1411 pKa = 3.7TSDD1414 pKa = 4.71LPAGYY1419 pKa = 9.72QLAANQPSSGTYY1431 pKa = 9.59TFTAEE1436 pKa = 4.24DD1437 pKa = 3.6NQVVQIHH1444 pKa = 5.48VVHH1447 pKa = 6.58IHH1449 pKa = 5.38TQSEE1453 pKa = 4.78MSTKK1457 pKa = 10.06DD1458 pKa = 2.99TVNYY1462 pKa = 10.6YY1463 pKa = 9.89EE1464 pKa = 4.84LPQDD1468 pKa = 3.64KK1469 pKa = 10.94AEE1471 pKa = 4.0QSKK1474 pKa = 8.83TEE1476 pKa = 4.09SVNWKK1481 pKa = 9.65IDD1483 pKa = 3.3TDD1485 pKa = 3.79EE1486 pKa = 4.25ATGISTFTPSSTAIEE1501 pKa = 4.1ISTPEE1506 pKa = 3.91VPGYY1510 pKa = 8.97TPSEE1514 pKa = 3.9QTIKK1518 pKa = 9.75FTQATSEE1525 pKa = 4.33TQPSDD1530 pKa = 3.0QTADD1534 pKa = 3.33VTYY1537 pKa = 10.65SADD1540 pKa = 3.46PQEE1543 pKa = 4.09ITFTYY1548 pKa = 10.82VDD1550 pKa = 4.25DD1551 pKa = 4.05TTGDD1555 pKa = 3.26EE1556 pKa = 4.53MEE1558 pKa = 4.84NLDD1561 pKa = 3.8ITLEE1565 pKa = 4.09GHH1567 pKa = 5.14TDD1569 pKa = 3.12EE1570 pKa = 5.76AGTYY1574 pKa = 7.63TVIVPKK1580 pKa = 10.63NYY1582 pKa = 9.97VLSQGQSATVPYY1594 pKa = 9.55TYY1596 pKa = 9.9TADD1599 pKa = 3.31VQTPTKK1605 pKa = 10.61VHH1607 pKa = 6.03LTHH1610 pKa = 7.34AIADD1614 pKa = 4.07GTVVTTRR1621 pKa = 11.84TITYY1625 pKa = 8.78VVNGNPTAAPKK1636 pKa = 10.67AVVQKK1641 pKa = 9.32ITWKK1645 pKa = 10.1VVRR1648 pKa = 11.84DD1649 pKa = 3.84MVTGTSYY1656 pKa = 10.23ATAQNGYY1663 pKa = 9.76DD1664 pKa = 3.46AVITPEE1670 pKa = 4.04LAGYY1674 pKa = 7.2TADD1677 pKa = 3.78KK1678 pKa = 11.04SGIAQEE1684 pKa = 3.93LLGRR1688 pKa = 11.84VDD1690 pKa = 3.6TADD1693 pKa = 3.96LADD1696 pKa = 3.72SAVTVTYY1703 pKa = 8.9TAIPVAPDD1711 pKa = 3.15NDD1713 pKa = 4.11SEE1715 pKa = 4.85NEE1717 pKa = 4.37VVTPTQPDD1725 pKa = 3.72TAVSGTMGSDD1735 pKa = 3.09VDD1737 pKa = 4.57NEE1739 pKa = 4.17QSDD1742 pKa = 4.19VTVLEE1747 pKa = 4.36KK1748 pKa = 11.02DD1749 pKa = 3.5NQKK1752 pKa = 8.37VTRR1755 pKa = 11.84DD1756 pKa = 3.64GNSDD1760 pKa = 3.42EE1761 pKa = 4.54TTKK1764 pKa = 11.22SNAFGSSAGIYY1775 pKa = 8.58VHH1777 pKa = 7.06GSTHH1781 pKa = 6.75ADD1783 pKa = 3.01AVPTVTTDD1791 pKa = 3.15DD1792 pKa = 3.71TGVKK1796 pKa = 10.02KK1797 pKa = 9.71GTANKK1802 pKa = 10.11VNITTAAKK1810 pKa = 10.33LPQTDD1815 pKa = 3.68EE1816 pKa = 4.22KK1817 pKa = 11.46QNGIWTIAGASLLGLFSLFGIGKK1840 pKa = 9.22KK1841 pKa = 9.5KK1842 pKa = 10.27RR1843 pKa = 11.84RR1844 pKa = 11.84EE1845 pKa = 3.71DD1846 pKa = 3.51DD1847 pKa = 3.16
Molecular weight: 195.32 kDa
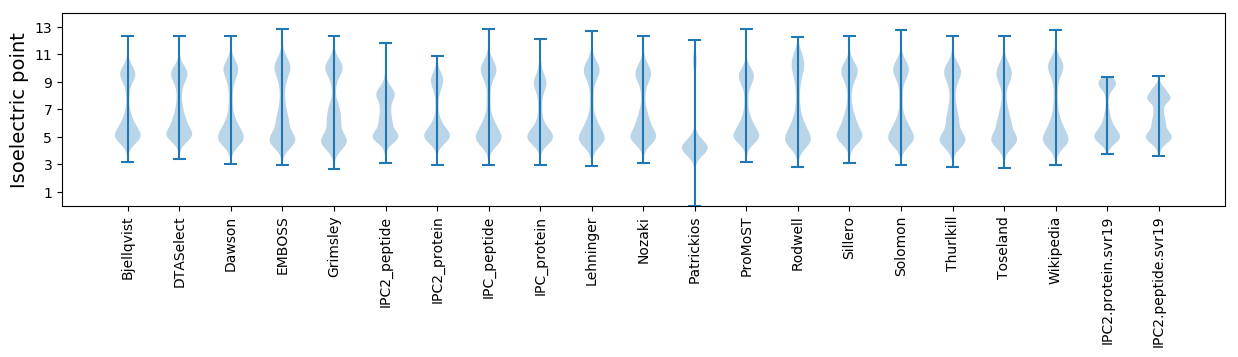
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R2B817|A0A0R2B817_LACCL Glycosyltransferase OS=Lactobacillus collinoides DSM 20515 = JCM 1123 OX=1423733 GN=FC82_GL002417 PE=4 SV=1
MM1 pKa = 7.55NILSQRR7 pKa = 11.84MIDD10 pKa = 3.61MSGHH14 pKa = 6.19RR15 pKa = 11.84IGRR18 pKa = 11.84LVVIKK23 pKa = 10.61RR24 pKa = 11.84AGKK27 pKa = 9.95LSNGNITWFCQCDD40 pKa = 3.57CGNQCVVDD48 pKa = 3.66GHH50 pKa = 6.27LLRR53 pKa = 11.84TGRR56 pKa = 11.84TLSCGCIRR64 pKa = 11.84KK65 pKa = 8.67QRR67 pKa = 11.84SRR69 pKa = 11.84EE70 pKa = 4.07LIRR73 pKa = 11.84NNPKK77 pKa = 8.26TRR79 pKa = 11.84QYY81 pKa = 10.43IGNWKK86 pKa = 9.07PLEE89 pKa = 4.38KK90 pKa = 9.12TWHH93 pKa = 6.04PRR95 pKa = 11.84ATDD98 pKa = 3.22RR99 pKa = 11.84RR100 pKa = 11.84KK101 pKa = 10.6NNSSGITGVSYY112 pKa = 10.27DD113 pKa = 3.72QKK115 pKa = 7.87QHH117 pKa = 5.08RR118 pKa = 11.84WIARR122 pKa = 11.84LYY124 pKa = 9.44FKK126 pKa = 10.81GRR128 pKa = 11.84YY129 pKa = 8.29VLNKK133 pKa = 9.83TFDD136 pKa = 3.56NKK138 pKa = 10.96ANAVEE143 pKa = 4.05ARR145 pKa = 11.84RR146 pKa = 11.84QAEE149 pKa = 4.25LRR151 pKa = 11.84FLTLQEE157 pKa = 4.59KK158 pKa = 10.7NEE160 pKa = 3.97QLL162 pKa = 3.5
MM1 pKa = 7.55NILSQRR7 pKa = 11.84MIDD10 pKa = 3.61MSGHH14 pKa = 6.19RR15 pKa = 11.84IGRR18 pKa = 11.84LVVIKK23 pKa = 10.61RR24 pKa = 11.84AGKK27 pKa = 9.95LSNGNITWFCQCDD40 pKa = 3.57CGNQCVVDD48 pKa = 3.66GHH50 pKa = 6.27LLRR53 pKa = 11.84TGRR56 pKa = 11.84TLSCGCIRR64 pKa = 11.84KK65 pKa = 8.67QRR67 pKa = 11.84SRR69 pKa = 11.84EE70 pKa = 4.07LIRR73 pKa = 11.84NNPKK77 pKa = 8.26TRR79 pKa = 11.84QYY81 pKa = 10.43IGNWKK86 pKa = 9.07PLEE89 pKa = 4.38KK90 pKa = 9.12TWHH93 pKa = 6.04PRR95 pKa = 11.84ATDD98 pKa = 3.22RR99 pKa = 11.84RR100 pKa = 11.84KK101 pKa = 10.6NNSSGITGVSYY112 pKa = 10.27DD113 pKa = 3.72QKK115 pKa = 7.87QHH117 pKa = 5.08RR118 pKa = 11.84WIARR122 pKa = 11.84LYY124 pKa = 9.44FKK126 pKa = 10.81GRR128 pKa = 11.84YY129 pKa = 8.29VLNKK133 pKa = 9.83TFDD136 pKa = 3.56NKK138 pKa = 10.96ANAVEE143 pKa = 4.05ARR145 pKa = 11.84RR146 pKa = 11.84QAEE149 pKa = 4.25LRR151 pKa = 11.84FLTLQEE157 pKa = 4.59KK158 pKa = 10.7NEE160 pKa = 3.97QLL162 pKa = 3.5
Molecular weight: 18.95 kDa
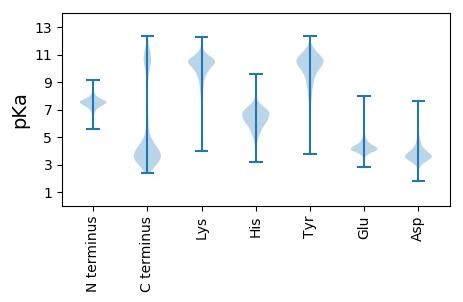
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
958480 |
50 |
3774 |
303.6 |
33.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.692 ± 0.047 | 0.485 ± 0.011 |
6.024 ± 0.044 | 4.87 ± 0.041 |
4.187 ± 0.031 | 6.919 ± 0.035 |
2.287 ± 0.023 | 6.701 ± 0.046 |
5.757 ± 0.045 | 9.7 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.691 ± 0.023 | 4.567 ± 0.034 |
3.705 ± 0.024 | 4.484 ± 0.037 |
3.972 ± 0.036 | 6.051 ± 0.049 |
7.039 ± 0.078 | 7.341 ± 0.037 |
1.069 ± 0.015 | 3.456 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
