
Kineobactrum sediminis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Kineobactrum
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
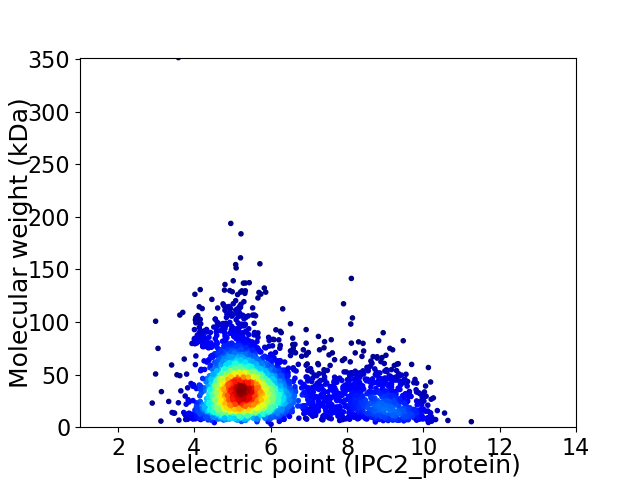
Virtual 2D-PAGE plot for 3544 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N5XZW2|A0A2N5XZW2_9GAMM LLM class F420-dependent oxidoreductase OS=Kineobactrum sediminis OX=1905677 GN=CWI75_14620 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 9.12VQHH5 pKa = 5.87VAVVCVVGLLSCLSQAAHH23 pKa = 6.74AAAPVAVEE31 pKa = 4.14DD32 pKa = 3.59NYY34 pKa = 11.57GRR36 pKa = 11.84FFSGANVLDD45 pKa = 4.12NDD47 pKa = 4.51SDD49 pKa = 4.86ADD51 pKa = 4.36DD52 pKa = 5.49DD53 pKa = 4.6PLSADD58 pKa = 4.94LVSQPQGGTVFLQTNGVFEE77 pKa = 4.13YY78 pKa = 10.6TPDD81 pKa = 3.53EE82 pKa = 4.78GFSGPDD88 pKa = 2.99TFQYY92 pKa = 10.23RR93 pKa = 11.84AEE95 pKa = 4.16AAGEE99 pKa = 3.77FSNVVNVYY107 pKa = 9.75LQVGGSTSEE116 pKa = 4.37SNTPPVAVDD125 pKa = 3.52DD126 pKa = 4.83NYY128 pKa = 11.59GGDD131 pKa = 3.62YY132 pKa = 10.71LGQNVLSNDD141 pKa = 3.08SDD143 pKa = 4.64AEE145 pKa = 4.2GDD147 pKa = 3.78PLAADD152 pKa = 4.9LVSQPLGGTVFMQLDD167 pKa = 4.05GTFSYY172 pKa = 10.27TPNEE176 pKa = 4.21GFSGSDD182 pKa = 2.88SFQYY186 pKa = 10.38RR187 pKa = 11.84AVANGDD193 pKa = 3.38FSNIATVTLFVEE205 pKa = 5.91GPANSPPVAQDD216 pKa = 2.98DD217 pKa = 4.94SYY219 pKa = 12.14GVDD222 pKa = 3.44YY223 pKa = 10.94QGKK226 pKa = 8.74NVLANDD232 pKa = 3.65TDD234 pKa = 3.74IDD236 pKa = 4.37EE237 pKa = 5.46DD238 pKa = 4.31PLTAILVTQPEE249 pKa = 4.94LGTVTLNSDD258 pKa = 3.46GTFTYY263 pKa = 10.01TPNTGASGNDD273 pKa = 2.85VFTYY277 pKa = 9.81QASDD281 pKa = 3.46GTDD284 pKa = 3.24LSNVATVSLLVEE296 pKa = 4.73APANSAPVAQDD307 pKa = 3.37DD308 pKa = 4.35NFGFDD313 pKa = 3.43YY314 pKa = 10.71QGRR317 pKa = 11.84NVLANDD323 pKa = 3.59TDD325 pKa = 3.69VDD327 pKa = 4.34EE328 pKa = 6.03DD329 pKa = 4.42PLTAILVTPIQNGSLTLNSDD349 pKa = 3.37GSFSYY354 pKa = 10.57TPATGFSGTDD364 pKa = 2.83SFTYY368 pKa = 9.72QASDD372 pKa = 3.17GSEE375 pKa = 4.02LSNTATVTLQVEE387 pKa = 4.87APAEE391 pKa = 4.17EE392 pKa = 4.44PVPLNPVALDD402 pKa = 3.91DD403 pKa = 4.6NFGADD408 pKa = 3.43FAGRR412 pKa = 11.84NVLANDD418 pKa = 4.15EE419 pKa = 4.68DD420 pKa = 4.46PSGAGLVALLEE431 pKa = 4.08QGPANGTVQLNSSGTFTYY449 pKa = 10.26EE450 pKa = 3.54PNAGFVGEE458 pKa = 5.07DD459 pKa = 3.37VFSYY463 pKa = 10.19RR464 pKa = 11.84ASNGEE469 pKa = 4.13LLSDD473 pKa = 3.81VATVTLLVVDD483 pKa = 4.3ATVTPAVAVDD493 pKa = 4.83DD494 pKa = 4.33EE495 pKa = 4.91FEE497 pKa = 5.21GDD499 pKa = 3.48FSGANLITNDD509 pKa = 4.57LYY511 pKa = 11.61ANPQDD516 pKa = 3.55LAAIKK521 pKa = 9.55VTDD524 pKa = 4.13PGEE527 pKa = 4.3GSLEE531 pKa = 4.07INPDD535 pKa = 2.72GSFTYY540 pKa = 10.35RR541 pKa = 11.84PFAGFTGNDD550 pKa = 2.95SFTYY554 pKa = 10.38LVRR557 pKa = 11.84DD558 pKa = 3.6PDD560 pKa = 3.95GLEE563 pKa = 4.24SNVATVLLTVDD574 pKa = 4.02GNTAGEE580 pKa = 4.38GGLAAKK586 pKa = 9.95KK587 pKa = 10.37DD588 pKa = 3.66ADD590 pKa = 3.97GTSVLVDD597 pKa = 3.44ATPNEE602 pKa = 3.99LAVARR607 pKa = 11.84RR608 pKa = 11.84FDD610 pKa = 5.09DD611 pKa = 3.13ICTRR615 pKa = 11.84LDD617 pKa = 3.57PANADD622 pKa = 3.67QQDD625 pKa = 4.48LLTLCANLRR634 pKa = 11.84KK635 pKa = 9.92QGTTAKK641 pKa = 9.98QALDD645 pKa = 3.76ALRR648 pKa = 11.84VITPEE653 pKa = 3.49EE654 pKa = 3.58LAAIGKK660 pKa = 5.66TVRR663 pKa = 11.84VLSFSQFRR671 pKa = 11.84NHH673 pKa = 6.25GSRR676 pKa = 11.84MARR679 pKa = 11.84VRR681 pKa = 11.84EE682 pKa = 4.0GSRR685 pKa = 11.84GVSLAGLDD693 pKa = 3.8LNYY696 pKa = 10.62EE697 pKa = 4.18NSRR700 pKa = 11.84VSGSTLDD707 pKa = 4.2ALLQEE712 pKa = 4.53SLGMGASGDD721 pKa = 3.59EE722 pKa = 4.04MLAGSRR728 pKa = 11.84LGVFVTGDD736 pKa = 3.87LSFGDD741 pKa = 3.55QDD743 pKa = 3.53RR744 pKa = 11.84TGLEE748 pKa = 3.83SGFDD752 pKa = 3.73FDD754 pKa = 5.18SQTLTVGMDD763 pKa = 3.21YY764 pKa = 11.12RR765 pKa = 11.84LTDD768 pKa = 3.37NAFVGTSLSIGQAEE782 pKa = 4.44VTFDD786 pKa = 3.42QQQGEE791 pKa = 4.69TTTDD795 pKa = 3.15NQALAVYY802 pKa = 10.01GSLYY806 pKa = 10.42SGNGYY811 pKa = 10.19VDD813 pKa = 5.59GIFSYY818 pKa = 10.36GWSDD822 pKa = 2.94VDD824 pKa = 3.44TEE826 pKa = 4.7RR827 pKa = 11.84NISYY831 pKa = 10.5QDD833 pKa = 3.07FGGSVEE839 pKa = 4.09RR840 pKa = 11.84TAAGSTDD847 pKa = 2.83GSEE850 pKa = 4.73YY851 pKa = 10.38YY852 pKa = 11.0VSVNAGYY859 pKa = 10.31SFSSGALRR867 pKa = 11.84LDD869 pKa = 3.13PVMRR873 pKa = 11.84FFYY876 pKa = 10.9LDD878 pKa = 3.31GSVDD882 pKa = 3.48GFTEE886 pKa = 4.4TGAQGLNLAVSDD898 pKa = 4.75NDD900 pKa = 3.83FQSMTVSASGQLSYY914 pKa = 10.79MFLPGWGVITPYY926 pKa = 10.8LRR928 pKa = 11.84LEE930 pKa = 4.14YY931 pKa = 10.16TRR933 pKa = 11.84EE934 pKa = 4.17LEE936 pKa = 4.18DD937 pKa = 3.05SAAGVRR943 pKa = 11.84YY944 pKa = 9.87RR945 pKa = 11.84FVNDD949 pKa = 3.96PFADD953 pKa = 3.37STLQIRR959 pKa = 11.84VDD961 pKa = 4.5DD962 pKa = 5.04PDD964 pKa = 3.41TSYY967 pKa = 10.61MVYY970 pKa = 10.29GAGVAAQFAHH980 pKa = 6.6GFSGFVSYY988 pKa = 10.51QALGGYY994 pKa = 9.98DD995 pKa = 3.58GLSGEE1000 pKa = 4.59TVSFGARR1007 pKa = 11.84WEE1009 pKa = 4.08LAFF1012 pKa = 5.37
MM1 pKa = 7.47KK2 pKa = 9.12VQHH5 pKa = 5.87VAVVCVVGLLSCLSQAAHH23 pKa = 6.74AAAPVAVEE31 pKa = 4.14DD32 pKa = 3.59NYY34 pKa = 11.57GRR36 pKa = 11.84FFSGANVLDD45 pKa = 4.12NDD47 pKa = 4.51SDD49 pKa = 4.86ADD51 pKa = 4.36DD52 pKa = 5.49DD53 pKa = 4.6PLSADD58 pKa = 4.94LVSQPQGGTVFLQTNGVFEE77 pKa = 4.13YY78 pKa = 10.6TPDD81 pKa = 3.53EE82 pKa = 4.78GFSGPDD88 pKa = 2.99TFQYY92 pKa = 10.23RR93 pKa = 11.84AEE95 pKa = 4.16AAGEE99 pKa = 3.77FSNVVNVYY107 pKa = 9.75LQVGGSTSEE116 pKa = 4.37SNTPPVAVDD125 pKa = 3.52DD126 pKa = 4.83NYY128 pKa = 11.59GGDD131 pKa = 3.62YY132 pKa = 10.71LGQNVLSNDD141 pKa = 3.08SDD143 pKa = 4.64AEE145 pKa = 4.2GDD147 pKa = 3.78PLAADD152 pKa = 4.9LVSQPLGGTVFMQLDD167 pKa = 4.05GTFSYY172 pKa = 10.27TPNEE176 pKa = 4.21GFSGSDD182 pKa = 2.88SFQYY186 pKa = 10.38RR187 pKa = 11.84AVANGDD193 pKa = 3.38FSNIATVTLFVEE205 pKa = 5.91GPANSPPVAQDD216 pKa = 2.98DD217 pKa = 4.94SYY219 pKa = 12.14GVDD222 pKa = 3.44YY223 pKa = 10.94QGKK226 pKa = 8.74NVLANDD232 pKa = 3.65TDD234 pKa = 3.74IDD236 pKa = 4.37EE237 pKa = 5.46DD238 pKa = 4.31PLTAILVTQPEE249 pKa = 4.94LGTVTLNSDD258 pKa = 3.46GTFTYY263 pKa = 10.01TPNTGASGNDD273 pKa = 2.85VFTYY277 pKa = 9.81QASDD281 pKa = 3.46GTDD284 pKa = 3.24LSNVATVSLLVEE296 pKa = 4.73APANSAPVAQDD307 pKa = 3.37DD308 pKa = 4.35NFGFDD313 pKa = 3.43YY314 pKa = 10.71QGRR317 pKa = 11.84NVLANDD323 pKa = 3.59TDD325 pKa = 3.69VDD327 pKa = 4.34EE328 pKa = 6.03DD329 pKa = 4.42PLTAILVTPIQNGSLTLNSDD349 pKa = 3.37GSFSYY354 pKa = 10.57TPATGFSGTDD364 pKa = 2.83SFTYY368 pKa = 9.72QASDD372 pKa = 3.17GSEE375 pKa = 4.02LSNTATVTLQVEE387 pKa = 4.87APAEE391 pKa = 4.17EE392 pKa = 4.44PVPLNPVALDD402 pKa = 3.91DD403 pKa = 4.6NFGADD408 pKa = 3.43FAGRR412 pKa = 11.84NVLANDD418 pKa = 4.15EE419 pKa = 4.68DD420 pKa = 4.46PSGAGLVALLEE431 pKa = 4.08QGPANGTVQLNSSGTFTYY449 pKa = 10.26EE450 pKa = 3.54PNAGFVGEE458 pKa = 5.07DD459 pKa = 3.37VFSYY463 pKa = 10.19RR464 pKa = 11.84ASNGEE469 pKa = 4.13LLSDD473 pKa = 3.81VATVTLLVVDD483 pKa = 4.3ATVTPAVAVDD493 pKa = 4.83DD494 pKa = 4.33EE495 pKa = 4.91FEE497 pKa = 5.21GDD499 pKa = 3.48FSGANLITNDD509 pKa = 4.57LYY511 pKa = 11.61ANPQDD516 pKa = 3.55LAAIKK521 pKa = 9.55VTDD524 pKa = 4.13PGEE527 pKa = 4.3GSLEE531 pKa = 4.07INPDD535 pKa = 2.72GSFTYY540 pKa = 10.35RR541 pKa = 11.84PFAGFTGNDD550 pKa = 2.95SFTYY554 pKa = 10.38LVRR557 pKa = 11.84DD558 pKa = 3.6PDD560 pKa = 3.95GLEE563 pKa = 4.24SNVATVLLTVDD574 pKa = 4.02GNTAGEE580 pKa = 4.38GGLAAKK586 pKa = 9.95KK587 pKa = 10.37DD588 pKa = 3.66ADD590 pKa = 3.97GTSVLVDD597 pKa = 3.44ATPNEE602 pKa = 3.99LAVARR607 pKa = 11.84RR608 pKa = 11.84FDD610 pKa = 5.09DD611 pKa = 3.13ICTRR615 pKa = 11.84LDD617 pKa = 3.57PANADD622 pKa = 3.67QQDD625 pKa = 4.48LLTLCANLRR634 pKa = 11.84KK635 pKa = 9.92QGTTAKK641 pKa = 9.98QALDD645 pKa = 3.76ALRR648 pKa = 11.84VITPEE653 pKa = 3.49EE654 pKa = 3.58LAAIGKK660 pKa = 5.66TVRR663 pKa = 11.84VLSFSQFRR671 pKa = 11.84NHH673 pKa = 6.25GSRR676 pKa = 11.84MARR679 pKa = 11.84VRR681 pKa = 11.84EE682 pKa = 4.0GSRR685 pKa = 11.84GVSLAGLDD693 pKa = 3.8LNYY696 pKa = 10.62EE697 pKa = 4.18NSRR700 pKa = 11.84VSGSTLDD707 pKa = 4.2ALLQEE712 pKa = 4.53SLGMGASGDD721 pKa = 3.59EE722 pKa = 4.04MLAGSRR728 pKa = 11.84LGVFVTGDD736 pKa = 3.87LSFGDD741 pKa = 3.55QDD743 pKa = 3.53RR744 pKa = 11.84TGLEE748 pKa = 3.83SGFDD752 pKa = 3.73FDD754 pKa = 5.18SQTLTVGMDD763 pKa = 3.21YY764 pKa = 11.12RR765 pKa = 11.84LTDD768 pKa = 3.37NAFVGTSLSIGQAEE782 pKa = 4.44VTFDD786 pKa = 3.42QQQGEE791 pKa = 4.69TTTDD795 pKa = 3.15NQALAVYY802 pKa = 10.01GSLYY806 pKa = 10.42SGNGYY811 pKa = 10.19VDD813 pKa = 5.59GIFSYY818 pKa = 10.36GWSDD822 pKa = 2.94VDD824 pKa = 3.44TEE826 pKa = 4.7RR827 pKa = 11.84NISYY831 pKa = 10.5QDD833 pKa = 3.07FGGSVEE839 pKa = 4.09RR840 pKa = 11.84TAAGSTDD847 pKa = 2.83GSEE850 pKa = 4.73YY851 pKa = 10.38YY852 pKa = 11.0VSVNAGYY859 pKa = 10.31SFSSGALRR867 pKa = 11.84LDD869 pKa = 3.13PVMRR873 pKa = 11.84FFYY876 pKa = 10.9LDD878 pKa = 3.31GSVDD882 pKa = 3.48GFTEE886 pKa = 4.4TGAQGLNLAVSDD898 pKa = 4.75NDD900 pKa = 3.83FQSMTVSASGQLSYY914 pKa = 10.79MFLPGWGVITPYY926 pKa = 10.8LRR928 pKa = 11.84LEE930 pKa = 4.14YY931 pKa = 10.16TRR933 pKa = 11.84EE934 pKa = 4.17LEE936 pKa = 4.18DD937 pKa = 3.05SAAGVRR943 pKa = 11.84YY944 pKa = 9.87RR945 pKa = 11.84FVNDD949 pKa = 3.96PFADD953 pKa = 3.37STLQIRR959 pKa = 11.84VDD961 pKa = 4.5DD962 pKa = 5.04PDD964 pKa = 3.41TSYY967 pKa = 10.61MVYY970 pKa = 10.29GAGVAAQFAHH980 pKa = 6.6GFSGFVSYY988 pKa = 10.51QALGGYY994 pKa = 9.98DD995 pKa = 3.58GLSGEE1000 pKa = 4.59TVSFGARR1007 pKa = 11.84WEE1009 pKa = 4.08LAFF1012 pKa = 5.37
Molecular weight: 106.65 kDa
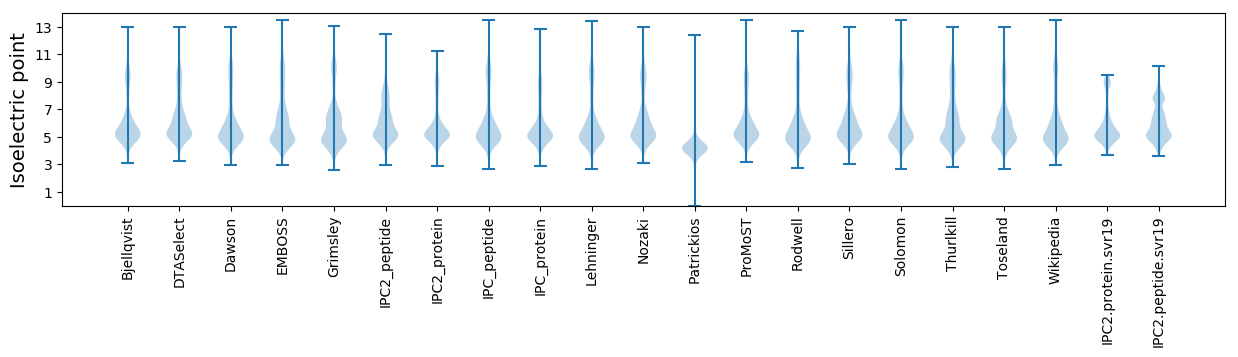
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N5Y544|A0A2N5Y544_9GAMM Type IV pili twitching motility protein PilT OS=Kineobactrum sediminis OX=1905677 GN=CWI75_03960 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.87RR14 pKa = 11.84THH16 pKa = 5.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.46NGRR28 pKa = 11.84LLLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.87RR14 pKa = 11.84THH16 pKa = 5.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.46NGRR28 pKa = 11.84LLLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LAAA44 pKa = 4.42
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1232078 |
27 |
3408 |
347.7 |
38.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.86 ± 0.046 | 1.029 ± 0.016 |
5.834 ± 0.035 | 6.164 ± 0.037 |
3.631 ± 0.027 | 8.171 ± 0.038 |
2.249 ± 0.022 | 5.05 ± 0.03 |
2.791 ± 0.032 | 10.861 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.384 ± 0.019 | 3.086 ± 0.026 |
4.845 ± 0.026 | 4.025 ± 0.028 |
6.711 ± 0.033 | 5.774 ± 0.026 |
5.217 ± 0.025 | 7.222 ± 0.034 |
1.399 ± 0.017 | 2.698 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
