
Faecalicatena orotica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Faecalicatena
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
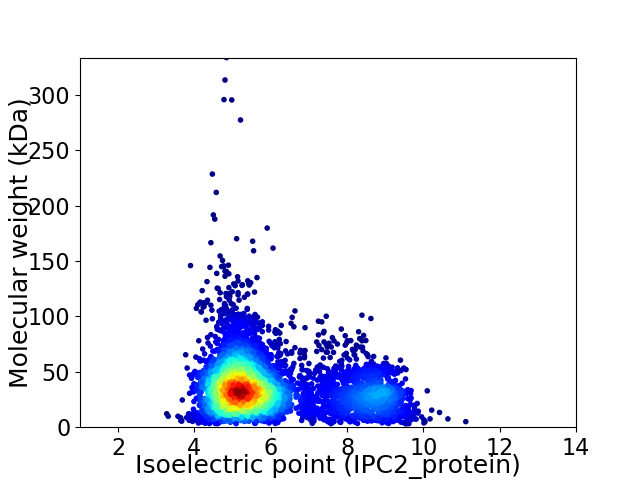
Virtual 2D-PAGE plot for 5138 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
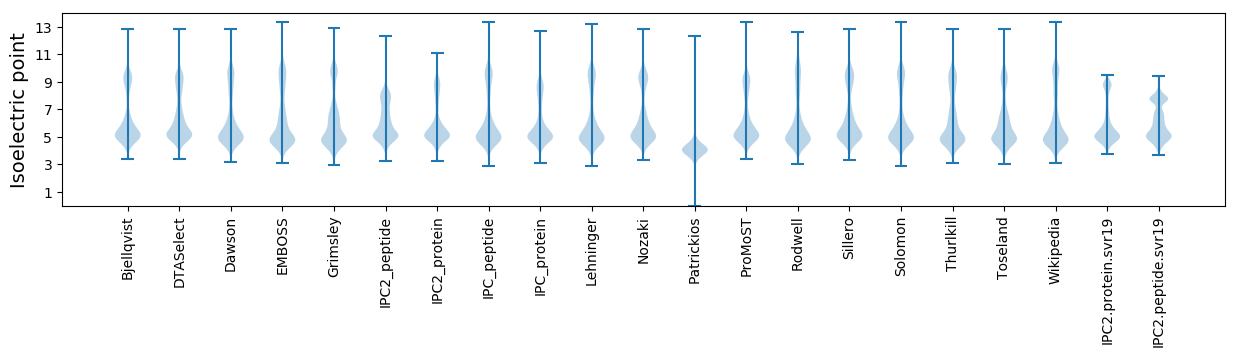
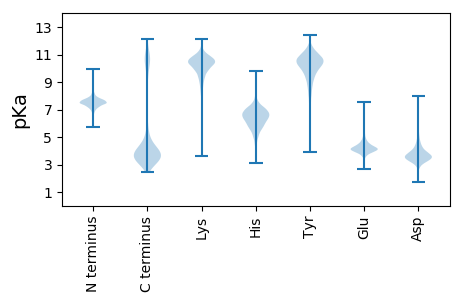
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Y9C692|A0A2Y9C692_9FIRM Formate acetyltransferase OS=Faecalicatena orotica OX=1544 GN=A8806_11473 PE=3 SV=1
MM1 pKa = 6.83STHH4 pKa = 5.59RR5 pKa = 11.84HH6 pKa = 5.32IDD8 pKa = 3.69KK9 pKa = 10.43ICCAVLLAVFILTAVFCNAEE29 pKa = 4.01ALGVEE34 pKa = 4.73TVSSAFGYY42 pKa = 9.84EE43 pKa = 3.85GKK45 pKa = 10.71LFDD48 pKa = 3.98TSSVHH53 pKa = 6.74TIDD56 pKa = 5.05IVMDD60 pKa = 3.69DD61 pKa = 3.12WDD63 pKa = 4.99GFLEE67 pKa = 4.35TCTNEE72 pKa = 3.88EE73 pKa = 4.12YY74 pKa = 10.36TICAVVIDD82 pKa = 3.83NEE84 pKa = 4.25AYY86 pKa = 9.58KK87 pKa = 10.94NVGIRR92 pKa = 11.84AKK94 pKa = 10.87GNTSLSSVAAYY105 pKa = 10.51GNDD108 pKa = 3.09RR109 pKa = 11.84YY110 pKa = 11.01SFKK113 pKa = 10.68IEE115 pKa = 3.57FDD117 pKa = 3.72HH118 pKa = 8.26YY119 pKa = 11.7DD120 pKa = 3.35DD121 pKa = 4.16AKK123 pKa = 11.21NYY125 pKa = 10.3YY126 pKa = 10.06GLDD129 pKa = 3.71KK130 pKa = 10.87ISLNNIIQDD139 pKa = 3.55NTYY142 pKa = 9.28MKK144 pKa = 10.48DD145 pKa = 3.53YY146 pKa = 10.92LSYY149 pKa = 11.58QMMGYY154 pKa = 10.18FGVDD158 pKa = 3.15APLCSYY164 pKa = 11.84ANITVNGEE172 pKa = 3.64DD173 pKa = 3.02WGLYY177 pKa = 9.44LAVEE181 pKa = 4.5GVEE184 pKa = 4.32EE185 pKa = 4.52SFLTRR190 pKa = 11.84NYY192 pKa = 10.49GSDD195 pKa = 3.3YY196 pKa = 11.43GEE198 pKa = 4.5LYY200 pKa = 10.7KK201 pKa = 10.38PDD203 pKa = 4.67SISMGGGRR211 pKa = 11.84GNGGGFDD218 pKa = 3.35MEE220 pKa = 4.66DD221 pKa = 3.39FEE223 pKa = 5.2EE224 pKa = 5.48RR225 pKa = 11.84SGQDD229 pKa = 3.07GEE231 pKa = 4.91ADD233 pKa = 3.59TEE235 pKa = 4.13EE236 pKa = 5.48DD237 pKa = 3.52SGKK240 pKa = 9.02DD241 pKa = 3.31TSGKK245 pKa = 8.24ATPGGGMKK253 pKa = 10.18PGGRR257 pKa = 11.84GQPEE261 pKa = 5.12DD262 pKa = 3.53GTLPDD267 pKa = 4.46MDD269 pKa = 5.58DD270 pKa = 3.84MPDD273 pKa = 3.27TGTMPEE279 pKa = 4.29KK280 pKa = 10.75GAMPDD285 pKa = 3.32MGDD288 pKa = 3.52MPEE291 pKa = 4.93DD292 pKa = 3.27GAMPDD297 pKa = 3.71MGNMPGDD304 pKa = 3.28GAMPDD309 pKa = 3.58MGDD312 pKa = 3.42MPEE315 pKa = 5.39DD316 pKa = 3.42GTLPEE321 pKa = 4.54MGDD324 pKa = 3.52MPGNGAMPANGGGMGMGSDD343 pKa = 4.34DD344 pKa = 3.47VSLIYY349 pKa = 10.62SDD351 pKa = 5.75DD352 pKa = 4.26DD353 pKa = 3.43QDD355 pKa = 3.98SYY357 pKa = 12.41SNIFDD362 pKa = 3.67NAKK365 pKa = 9.91TNVSDD370 pKa = 3.81ADD372 pKa = 3.57KK373 pKa = 10.96EE374 pKa = 4.13RR375 pKa = 11.84LIEE378 pKa = 4.17SLKK381 pKa = 9.7TLNEE385 pKa = 4.23GGDD388 pKa = 3.46IEE390 pKa = 5.21SVVDD394 pKa = 3.33VDD396 pKa = 3.45EE397 pKa = 4.76VIRR400 pKa = 11.84YY401 pKa = 7.5FVVHH405 pKa = 6.34NFVCNFDD412 pKa = 4.16SYY414 pKa = 9.87TGSMIHH420 pKa = 6.56NYY422 pKa = 9.82YY423 pKa = 10.43LYY425 pKa = 10.83EE426 pKa = 4.36EE427 pKa = 5.03DD428 pKa = 5.07GVMSMIPWDD437 pKa = 3.65YY438 pKa = 11.35NLAFGGFMSGSDD450 pKa = 3.24AGSLVNYY457 pKa = 9.61PIDD460 pKa = 3.82TPVSGGTVEE469 pKa = 4.23SRR471 pKa = 11.84PMLAWIFNEE480 pKa = 3.84EE481 pKa = 4.58AYY483 pKa = 9.94TEE485 pKa = 4.33IYY487 pKa = 10.12HH488 pKa = 6.82EE489 pKa = 4.33YY490 pKa = 8.54FAEE493 pKa = 5.19FISEE497 pKa = 4.28YY498 pKa = 10.23FDD500 pKa = 3.05SGYY503 pKa = 10.0FEE505 pKa = 5.35EE506 pKa = 5.37MIDD509 pKa = 4.16SVSEE513 pKa = 3.87MIAPYY518 pKa = 10.51VEE520 pKa = 4.0NDD522 pKa = 3.19PTKK525 pKa = 9.44FCTYY529 pKa = 10.55EE530 pKa = 3.81EE531 pKa = 4.4FEE533 pKa = 4.7EE534 pKa = 5.21GVSTLKK540 pKa = 10.53EE541 pKa = 3.98FCLLRR546 pKa = 11.84AEE548 pKa = 4.93SINGQLDD555 pKa = 3.45GTIPSTSDD563 pKa = 3.04GQGEE567 pKa = 4.43DD568 pKa = 3.76DD569 pKa = 4.9SALIDD574 pKa = 3.93AGDD577 pKa = 3.86IEE579 pKa = 4.65VSAMGSMGNMMGMGGSKK596 pKa = 10.55NKK598 pKa = 9.72GRR600 pKa = 4.06
MM1 pKa = 6.83STHH4 pKa = 5.59RR5 pKa = 11.84HH6 pKa = 5.32IDD8 pKa = 3.69KK9 pKa = 10.43ICCAVLLAVFILTAVFCNAEE29 pKa = 4.01ALGVEE34 pKa = 4.73TVSSAFGYY42 pKa = 9.84EE43 pKa = 3.85GKK45 pKa = 10.71LFDD48 pKa = 3.98TSSVHH53 pKa = 6.74TIDD56 pKa = 5.05IVMDD60 pKa = 3.69DD61 pKa = 3.12WDD63 pKa = 4.99GFLEE67 pKa = 4.35TCTNEE72 pKa = 3.88EE73 pKa = 4.12YY74 pKa = 10.36TICAVVIDD82 pKa = 3.83NEE84 pKa = 4.25AYY86 pKa = 9.58KK87 pKa = 10.94NVGIRR92 pKa = 11.84AKK94 pKa = 10.87GNTSLSSVAAYY105 pKa = 10.51GNDD108 pKa = 3.09RR109 pKa = 11.84YY110 pKa = 11.01SFKK113 pKa = 10.68IEE115 pKa = 3.57FDD117 pKa = 3.72HH118 pKa = 8.26YY119 pKa = 11.7DD120 pKa = 3.35DD121 pKa = 4.16AKK123 pKa = 11.21NYY125 pKa = 10.3YY126 pKa = 10.06GLDD129 pKa = 3.71KK130 pKa = 10.87ISLNNIIQDD139 pKa = 3.55NTYY142 pKa = 9.28MKK144 pKa = 10.48DD145 pKa = 3.53YY146 pKa = 10.92LSYY149 pKa = 11.58QMMGYY154 pKa = 10.18FGVDD158 pKa = 3.15APLCSYY164 pKa = 11.84ANITVNGEE172 pKa = 3.64DD173 pKa = 3.02WGLYY177 pKa = 9.44LAVEE181 pKa = 4.5GVEE184 pKa = 4.32EE185 pKa = 4.52SFLTRR190 pKa = 11.84NYY192 pKa = 10.49GSDD195 pKa = 3.3YY196 pKa = 11.43GEE198 pKa = 4.5LYY200 pKa = 10.7KK201 pKa = 10.38PDD203 pKa = 4.67SISMGGGRR211 pKa = 11.84GNGGGFDD218 pKa = 3.35MEE220 pKa = 4.66DD221 pKa = 3.39FEE223 pKa = 5.2EE224 pKa = 5.48RR225 pKa = 11.84SGQDD229 pKa = 3.07GEE231 pKa = 4.91ADD233 pKa = 3.59TEE235 pKa = 4.13EE236 pKa = 5.48DD237 pKa = 3.52SGKK240 pKa = 9.02DD241 pKa = 3.31TSGKK245 pKa = 8.24ATPGGGMKK253 pKa = 10.18PGGRR257 pKa = 11.84GQPEE261 pKa = 5.12DD262 pKa = 3.53GTLPDD267 pKa = 4.46MDD269 pKa = 5.58DD270 pKa = 3.84MPDD273 pKa = 3.27TGTMPEE279 pKa = 4.29KK280 pKa = 10.75GAMPDD285 pKa = 3.32MGDD288 pKa = 3.52MPEE291 pKa = 4.93DD292 pKa = 3.27GAMPDD297 pKa = 3.71MGNMPGDD304 pKa = 3.28GAMPDD309 pKa = 3.58MGDD312 pKa = 3.42MPEE315 pKa = 5.39DD316 pKa = 3.42GTLPEE321 pKa = 4.54MGDD324 pKa = 3.52MPGNGAMPANGGGMGMGSDD343 pKa = 4.34DD344 pKa = 3.47VSLIYY349 pKa = 10.62SDD351 pKa = 5.75DD352 pKa = 4.26DD353 pKa = 3.43QDD355 pKa = 3.98SYY357 pKa = 12.41SNIFDD362 pKa = 3.67NAKK365 pKa = 9.91TNVSDD370 pKa = 3.81ADD372 pKa = 3.57KK373 pKa = 10.96EE374 pKa = 4.13RR375 pKa = 11.84LIEE378 pKa = 4.17SLKK381 pKa = 9.7TLNEE385 pKa = 4.23GGDD388 pKa = 3.46IEE390 pKa = 5.21SVVDD394 pKa = 3.33VDD396 pKa = 3.45EE397 pKa = 4.76VIRR400 pKa = 11.84YY401 pKa = 7.5FVVHH405 pKa = 6.34NFVCNFDD412 pKa = 4.16SYY414 pKa = 9.87TGSMIHH420 pKa = 6.56NYY422 pKa = 9.82YY423 pKa = 10.43LYY425 pKa = 10.83EE426 pKa = 4.36EE427 pKa = 5.03DD428 pKa = 5.07GVMSMIPWDD437 pKa = 3.65YY438 pKa = 11.35NLAFGGFMSGSDD450 pKa = 3.24AGSLVNYY457 pKa = 9.61PIDD460 pKa = 3.82TPVSGGTVEE469 pKa = 4.23SRR471 pKa = 11.84PMLAWIFNEE480 pKa = 3.84EE481 pKa = 4.58AYY483 pKa = 9.94TEE485 pKa = 4.33IYY487 pKa = 10.12HH488 pKa = 6.82EE489 pKa = 4.33YY490 pKa = 8.54FAEE493 pKa = 5.19FISEE497 pKa = 4.28YY498 pKa = 10.23FDD500 pKa = 3.05SGYY503 pKa = 10.0FEE505 pKa = 5.35EE506 pKa = 5.37MIDD509 pKa = 4.16SVSEE513 pKa = 3.87MIAPYY518 pKa = 10.51VEE520 pKa = 4.0NDD522 pKa = 3.19PTKK525 pKa = 9.44FCTYY529 pKa = 10.55EE530 pKa = 3.81EE531 pKa = 4.4FEE533 pKa = 4.7EE534 pKa = 5.21GVSTLKK540 pKa = 10.53EE541 pKa = 3.98FCLLRR546 pKa = 11.84AEE548 pKa = 4.93SINGQLDD555 pKa = 3.45GTIPSTSDD563 pKa = 3.04GQGEE567 pKa = 4.43DD568 pKa = 3.76DD569 pKa = 4.9SALIDD574 pKa = 3.93AGDD577 pKa = 3.86IEE579 pKa = 4.65VSAMGSMGNMMGMGGSKK596 pKa = 10.55NKK598 pKa = 9.72GRR600 pKa = 4.06
Molecular weight: 65.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Y9CAT9|A0A2Y9CAT9_9FIRM Protein N-acetyltransferase RimJ/RimL family OS=Faecalicatena orotica OX=1544 GN=A8806_12310 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.73MTFQPKK8 pKa = 8.63NRR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.89VHH16 pKa = 5.88GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.73MTFQPKK8 pKa = 8.63NRR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.89VHH16 pKa = 5.88GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1715350 |
28 |
2958 |
333.9 |
37.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.391 ± 0.035 | 1.523 ± 0.014 |
5.585 ± 0.023 | 7.671 ± 0.037 |
4.197 ± 0.022 | 7.317 ± 0.029 |
1.753 ± 0.013 | 7.437 ± 0.033 |
6.656 ± 0.025 | 9.02 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.245 ± 0.019 | 4.19 ± 0.022 |
3.349 ± 0.02 | 3.218 ± 0.019 |
4.335 ± 0.022 | 5.761 ± 0.025 |
5.292 ± 0.025 | 6.878 ± 0.027 |
0.967 ± 0.012 | 4.215 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
