
bacterium D16-50
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
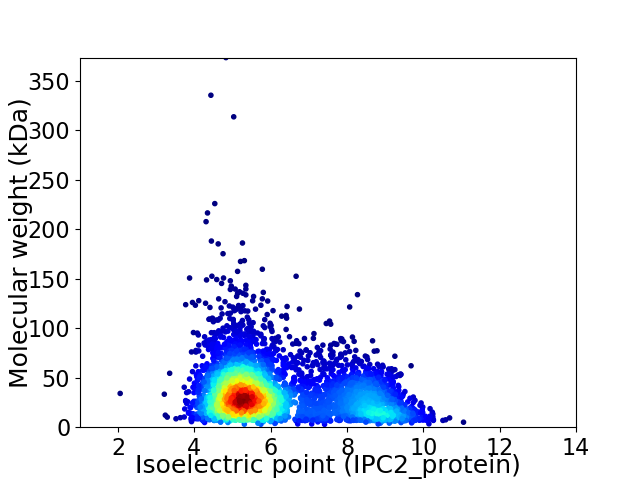
Virtual 2D-PAGE plot for 4338 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A9D3L7|A0A3A9D3L7_9BACT Esterase family protein OS=bacterium D16-50 OX=2320112 GN=D7X48_18660 PE=4 SV=1
MM1 pKa = 7.3KK2 pKa = 10.35RR3 pKa = 11.84KK4 pKa = 9.49LLSLVLSAALVLSMAACGNEE24 pKa = 3.92DD25 pKa = 3.52EE26 pKa = 5.04PANGSTPQDD35 pKa = 3.6SVSLSSEE42 pKa = 4.31PEE44 pKa = 3.84SSSAGEE50 pKa = 4.0PSDD53 pKa = 5.01EE54 pKa = 4.5GDD56 pKa = 3.71ASDD59 pKa = 4.44ASDD62 pKa = 3.29VGGAASGEE70 pKa = 4.01PTVIRR75 pKa = 11.84FGTHH79 pKa = 5.01WVPEE83 pKa = 4.26LDD85 pKa = 3.41PYY87 pKa = 9.96YY88 pKa = 9.75TDD90 pKa = 3.77EE91 pKa = 4.28VTGEE95 pKa = 3.87YY96 pKa = 10.77TMGEE100 pKa = 4.36RR101 pKa = 11.84EE102 pKa = 4.04RR103 pKa = 11.84QAALAGLEE111 pKa = 4.42AIKK114 pKa = 10.4KK115 pKa = 8.77AYY117 pKa = 9.68NVEE120 pKa = 4.5FEE122 pKa = 4.11FLQYY126 pKa = 10.44PVDD129 pKa = 3.85VASDD133 pKa = 3.6LMTSVLAQDD142 pKa = 5.27PICDD146 pKa = 3.99LALMWGGVEE155 pKa = 4.02PTILAQNVLQDD166 pKa = 3.4LSAYY170 pKa = 9.0TGLFEE175 pKa = 6.52DD176 pKa = 6.13DD177 pKa = 3.41EE178 pKa = 5.21ASWLLKK184 pKa = 10.6GSVFDD189 pKa = 4.62GYY191 pKa = 11.73YY192 pKa = 10.37LLGYY196 pKa = 9.87EE197 pKa = 4.5FGEE200 pKa = 4.44TSFPLIVNMTMLEE213 pKa = 4.15AVDD216 pKa = 3.89SLKK219 pKa = 11.19DD220 pKa = 3.73EE221 pKa = 4.5NGKK224 pKa = 8.31TIYY227 pKa = 8.85PTDD230 pKa = 3.64LFKK233 pKa = 10.96EE234 pKa = 5.01GKK236 pKa = 6.35WTWSTFKK243 pKa = 10.93DD244 pKa = 3.51YY245 pKa = 10.85LAKK248 pKa = 10.43IKK250 pKa = 10.41AYY252 pKa = 8.9YY253 pKa = 10.4SNVAAPDD260 pKa = 3.54GAYY263 pKa = 10.4YY264 pKa = 10.51DD265 pKa = 3.95YY266 pKa = 11.27VQAYY270 pKa = 6.54EE271 pKa = 3.84TDD273 pKa = 3.51FRR275 pKa = 11.84YY276 pKa = 9.89AALGAVHH283 pKa = 7.03ANGGAIYY290 pKa = 10.27DD291 pKa = 3.97GGVTADD297 pKa = 3.24SAEE300 pKa = 4.26TIEE303 pKa = 4.09AVAYY307 pKa = 9.43IQEE310 pKa = 4.36LVDD313 pKa = 4.19EE314 pKa = 4.78EE315 pKa = 5.41LLTACHH321 pKa = 6.48LQDD324 pKa = 5.37DD325 pKa = 4.82YY326 pKa = 11.68TPEE329 pKa = 3.97WLRR332 pKa = 11.84GGNDD336 pKa = 3.17FGKK339 pKa = 10.49GATVFTDD346 pKa = 3.22CGGWMIGGHH355 pKa = 4.91STEE358 pKa = 4.28CAARR362 pKa = 11.84GEE364 pKa = 4.64SIGIIPWPMPDD375 pKa = 3.84DD376 pKa = 3.61ATVDD380 pKa = 3.63SEE382 pKa = 4.92TYY384 pKa = 9.8RR385 pKa = 11.84QSTNGGNSVGVLKK398 pKa = 10.75GVSPEE403 pKa = 3.98KK404 pKa = 10.48TEE406 pKa = 4.14LALKK410 pKa = 10.75AFILYY415 pKa = 7.24WQTYY419 pKa = 7.91HH420 pKa = 6.07KK421 pKa = 9.31TIAGVDD427 pKa = 4.05TIADD431 pKa = 3.56YY432 pKa = 10.62HH433 pKa = 6.63AAAALDD439 pKa = 3.6KK440 pKa = 10.84LAGYY444 pKa = 10.1GVDD447 pKa = 5.41LYY449 pKa = 11.58NEE451 pKa = 4.52TYY453 pKa = 11.03GDD455 pKa = 4.44DD456 pKa = 5.93LIDD459 pKa = 4.4CYY461 pKa = 10.63TYY463 pKa = 11.03IMSHH467 pKa = 6.09MSTNYY472 pKa = 10.49ANMMGLWEE480 pKa = 5.64DD481 pKa = 4.56YY482 pKa = 8.13NTHH485 pKa = 5.96DD486 pKa = 3.4QWTEE490 pKa = 3.4ILGQSLFKK498 pKa = 10.98APGMSSYY505 pKa = 11.43DD506 pKa = 3.45VAIKK510 pKa = 10.92ANLTNLTNKK519 pKa = 8.73TDD521 pKa = 3.77SIAAVLKK528 pKa = 10.32TEE530 pKa = 4.5GVHH533 pKa = 7.47DD534 pKa = 4.0NQKK537 pKa = 10.95PNITKK542 pKa = 10.24EE543 pKa = 4.04KK544 pKa = 10.59AILTAGAADD553 pKa = 3.99VDD555 pKa = 3.6WTQYY559 pKa = 10.45FSAEE563 pKa = 4.1DD564 pKa = 3.5AVDD567 pKa = 3.92GVIEE571 pKa = 4.1ITADD575 pKa = 3.44SITVNEE581 pKa = 4.35GLDD584 pKa = 3.59LNTPGKK590 pKa = 9.76YY591 pKa = 9.42EE592 pKa = 3.89KK593 pKa = 10.77AVTAKK598 pKa = 10.85ASDD601 pKa = 3.31KK602 pKa = 11.09SGNEE606 pKa = 3.65ASSDD610 pKa = 3.53LTVIVYY616 pKa = 10.1DD617 pKa = 4.89ADD619 pKa = 3.66NTDD622 pKa = 3.73APTAAAKK629 pKa = 10.63EE630 pKa = 4.01EE631 pKa = 4.4LPTVALNTEE640 pKa = 3.99TSGINWSDD648 pKa = 3.52FLEE651 pKa = 4.5SAEE654 pKa = 4.59DD655 pKa = 3.83ADD657 pKa = 4.94GVDD660 pKa = 3.41VKK662 pKa = 11.72DD663 pKa = 4.27NVTADD668 pKa = 4.06LSTLDD673 pKa = 3.31TTTPGEE679 pKa = 4.15YY680 pKa = 10.27DD681 pKa = 3.37VTLTVTDD688 pKa = 3.93YY689 pKa = 11.45VGNTAEE695 pKa = 4.13ITVKK699 pKa = 9.43VTVVSEE705 pKa = 4.05
MM1 pKa = 7.3KK2 pKa = 10.35RR3 pKa = 11.84KK4 pKa = 9.49LLSLVLSAALVLSMAACGNEE24 pKa = 3.92DD25 pKa = 3.52EE26 pKa = 5.04PANGSTPQDD35 pKa = 3.6SVSLSSEE42 pKa = 4.31PEE44 pKa = 3.84SSSAGEE50 pKa = 4.0PSDD53 pKa = 5.01EE54 pKa = 4.5GDD56 pKa = 3.71ASDD59 pKa = 4.44ASDD62 pKa = 3.29VGGAASGEE70 pKa = 4.01PTVIRR75 pKa = 11.84FGTHH79 pKa = 5.01WVPEE83 pKa = 4.26LDD85 pKa = 3.41PYY87 pKa = 9.96YY88 pKa = 9.75TDD90 pKa = 3.77EE91 pKa = 4.28VTGEE95 pKa = 3.87YY96 pKa = 10.77TMGEE100 pKa = 4.36RR101 pKa = 11.84EE102 pKa = 4.04RR103 pKa = 11.84QAALAGLEE111 pKa = 4.42AIKK114 pKa = 10.4KK115 pKa = 8.77AYY117 pKa = 9.68NVEE120 pKa = 4.5FEE122 pKa = 4.11FLQYY126 pKa = 10.44PVDD129 pKa = 3.85VASDD133 pKa = 3.6LMTSVLAQDD142 pKa = 5.27PICDD146 pKa = 3.99LALMWGGVEE155 pKa = 4.02PTILAQNVLQDD166 pKa = 3.4LSAYY170 pKa = 9.0TGLFEE175 pKa = 6.52DD176 pKa = 6.13DD177 pKa = 3.41EE178 pKa = 5.21ASWLLKK184 pKa = 10.6GSVFDD189 pKa = 4.62GYY191 pKa = 11.73YY192 pKa = 10.37LLGYY196 pKa = 9.87EE197 pKa = 4.5FGEE200 pKa = 4.44TSFPLIVNMTMLEE213 pKa = 4.15AVDD216 pKa = 3.89SLKK219 pKa = 11.19DD220 pKa = 3.73EE221 pKa = 4.5NGKK224 pKa = 8.31TIYY227 pKa = 8.85PTDD230 pKa = 3.64LFKK233 pKa = 10.96EE234 pKa = 5.01GKK236 pKa = 6.35WTWSTFKK243 pKa = 10.93DD244 pKa = 3.51YY245 pKa = 10.85LAKK248 pKa = 10.43IKK250 pKa = 10.41AYY252 pKa = 8.9YY253 pKa = 10.4SNVAAPDD260 pKa = 3.54GAYY263 pKa = 10.4YY264 pKa = 10.51DD265 pKa = 3.95YY266 pKa = 11.27VQAYY270 pKa = 6.54EE271 pKa = 3.84TDD273 pKa = 3.51FRR275 pKa = 11.84YY276 pKa = 9.89AALGAVHH283 pKa = 7.03ANGGAIYY290 pKa = 10.27DD291 pKa = 3.97GGVTADD297 pKa = 3.24SAEE300 pKa = 4.26TIEE303 pKa = 4.09AVAYY307 pKa = 9.43IQEE310 pKa = 4.36LVDD313 pKa = 4.19EE314 pKa = 4.78EE315 pKa = 5.41LLTACHH321 pKa = 6.48LQDD324 pKa = 5.37DD325 pKa = 4.82YY326 pKa = 11.68TPEE329 pKa = 3.97WLRR332 pKa = 11.84GGNDD336 pKa = 3.17FGKK339 pKa = 10.49GATVFTDD346 pKa = 3.22CGGWMIGGHH355 pKa = 4.91STEE358 pKa = 4.28CAARR362 pKa = 11.84GEE364 pKa = 4.64SIGIIPWPMPDD375 pKa = 3.84DD376 pKa = 3.61ATVDD380 pKa = 3.63SEE382 pKa = 4.92TYY384 pKa = 9.8RR385 pKa = 11.84QSTNGGNSVGVLKK398 pKa = 10.75GVSPEE403 pKa = 3.98KK404 pKa = 10.48TEE406 pKa = 4.14LALKK410 pKa = 10.75AFILYY415 pKa = 7.24WQTYY419 pKa = 7.91HH420 pKa = 6.07KK421 pKa = 9.31TIAGVDD427 pKa = 4.05TIADD431 pKa = 3.56YY432 pKa = 10.62HH433 pKa = 6.63AAAALDD439 pKa = 3.6KK440 pKa = 10.84LAGYY444 pKa = 10.1GVDD447 pKa = 5.41LYY449 pKa = 11.58NEE451 pKa = 4.52TYY453 pKa = 11.03GDD455 pKa = 4.44DD456 pKa = 5.93LIDD459 pKa = 4.4CYY461 pKa = 10.63TYY463 pKa = 11.03IMSHH467 pKa = 6.09MSTNYY472 pKa = 10.49ANMMGLWEE480 pKa = 5.64DD481 pKa = 4.56YY482 pKa = 8.13NTHH485 pKa = 5.96DD486 pKa = 3.4QWTEE490 pKa = 3.4ILGQSLFKK498 pKa = 10.98APGMSSYY505 pKa = 11.43DD506 pKa = 3.45VAIKK510 pKa = 10.92ANLTNLTNKK519 pKa = 8.73TDD521 pKa = 3.77SIAAVLKK528 pKa = 10.32TEE530 pKa = 4.5GVHH533 pKa = 7.47DD534 pKa = 4.0NQKK537 pKa = 10.95PNITKK542 pKa = 10.24EE543 pKa = 4.04KK544 pKa = 10.59AILTAGAADD553 pKa = 3.99VDD555 pKa = 3.6WTQYY559 pKa = 10.45FSAEE563 pKa = 4.1DD564 pKa = 3.5AVDD567 pKa = 3.92GVIEE571 pKa = 4.1ITADD575 pKa = 3.44SITVNEE581 pKa = 4.35GLDD584 pKa = 3.59LNTPGKK590 pKa = 9.76YY591 pKa = 9.42EE592 pKa = 3.89KK593 pKa = 10.77AVTAKK598 pKa = 10.85ASDD601 pKa = 3.31KK602 pKa = 11.09SGNEE606 pKa = 3.65ASSDD610 pKa = 3.53LTVIVYY616 pKa = 10.1DD617 pKa = 4.89ADD619 pKa = 3.66NTDD622 pKa = 3.73APTAAAKK629 pKa = 10.63EE630 pKa = 4.01EE631 pKa = 4.4LPTVALNTEE640 pKa = 3.99TSGINWSDD648 pKa = 3.52FLEE651 pKa = 4.5SAEE654 pKa = 4.59DD655 pKa = 3.83ADD657 pKa = 4.94GVDD660 pKa = 3.41VKK662 pKa = 11.72DD663 pKa = 4.27NVTADD668 pKa = 4.06LSTLDD673 pKa = 3.31TTTPGEE679 pKa = 4.15YY680 pKa = 10.27DD681 pKa = 3.37VTLTVTDD688 pKa = 3.93YY689 pKa = 11.45VGNTAEE695 pKa = 4.13ITVKK699 pKa = 9.43VTVVSEE705 pKa = 4.05
Molecular weight: 76.07 kDa
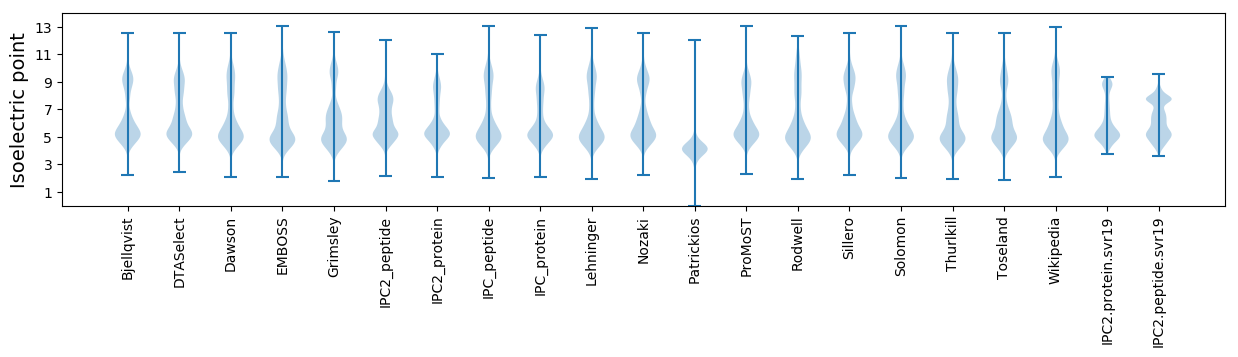
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9CZ26|A0A3A9CZ26_9BACT Uncharacterized protein OS=bacterium D16-50 OX=2320112 GN=D7X48_20390 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.58QPKK8 pKa = 9.19KK9 pKa = 8.22RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTRR25 pKa = 11.84GGRR28 pKa = 11.84KK29 pKa = 8.61VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84KK40 pKa = 8.68ILSAA44 pKa = 4.02
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.58QPKK8 pKa = 9.19KK9 pKa = 8.22RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTRR25 pKa = 11.84GGRR28 pKa = 11.84KK29 pKa = 8.61VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84KK40 pKa = 8.68ILSAA44 pKa = 4.02
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1406950 |
25 |
3304 |
324.3 |
36.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.936 ± 0.04 | 1.589 ± 0.015 |
5.505 ± 0.028 | 8.003 ± 0.046 |
4.037 ± 0.026 | 7.831 ± 0.044 |
1.701 ± 0.017 | 6.303 ± 0.04 |
5.991 ± 0.04 | 9.29 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.081 ± 0.017 | 3.857 ± 0.026 |
3.401 ± 0.024 | 3.421 ± 0.024 |
5.553 ± 0.038 | 5.749 ± 0.034 |
4.829 ± 0.035 | 6.629 ± 0.032 |
1.076 ± 0.016 | 4.217 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
