
Bizionia paragorgiae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Bizionia
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
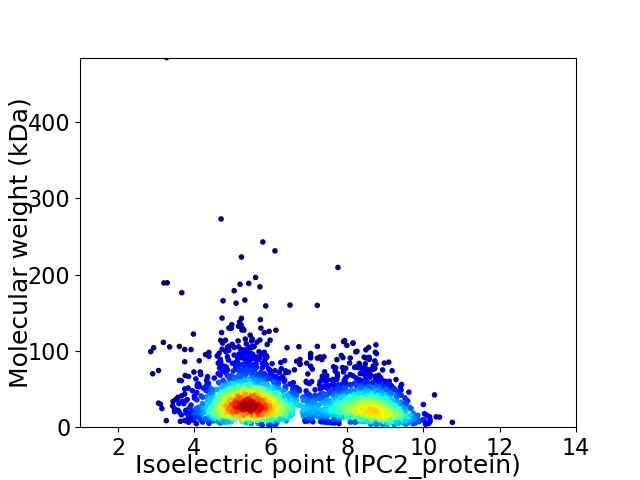
Virtual 2D-PAGE plot for 2859 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3YW71|A0A1H3YW71_9FLAO Cystathionine gamma-lyase OS=Bizionia paragorgiae OX=283786 GN=SAMN04487990_107113 PE=3 SV=1
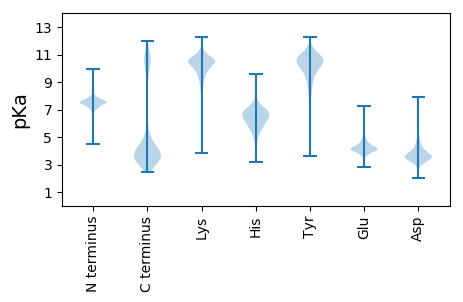
MM1 pKa = 7.17KK2 pKa = 9.86TNKK5 pKa = 9.65FLTLILSFVAVFAMTSCVEE24 pKa = 4.48DD25 pKa = 4.45DD26 pKa = 4.72DD27 pKa = 4.46FTVPSSLGNEE37 pKa = 3.78EE38 pKa = 3.65NAALVEE44 pKa = 4.0LLEE47 pKa = 4.56TSTEE51 pKa = 3.35VDD53 pKa = 2.95MAYY56 pKa = 10.57VQGLYY61 pKa = 10.25TSGTAPEE68 pKa = 4.44QIAADD73 pKa = 4.05IYY75 pKa = 9.63VKK77 pKa = 10.72GYY79 pKa = 10.41VSSSDD84 pKa = 3.2ATGNFYY90 pKa = 11.0KK91 pKa = 10.25EE92 pKa = 5.23FYY94 pKa = 9.55MQDD97 pKa = 3.27TPSNPTKK104 pKa = 10.19ALKK107 pKa = 10.34IIVSQSDD114 pKa = 3.89TYY116 pKa = 11.53NQFNKK121 pKa = 9.74GRR123 pKa = 11.84EE124 pKa = 3.93VYY126 pKa = 10.44INLKK130 pKa = 9.33GLYY133 pKa = 9.51VGEE136 pKa = 4.03EE137 pKa = 3.74RR138 pKa = 11.84VGNGVYY144 pKa = 10.21TIGGSVEE151 pKa = 3.77TDD153 pKa = 3.01QFGTTVQQLDD163 pKa = 3.48EE164 pKa = 4.72HH165 pKa = 6.08QVEE168 pKa = 4.66TKK170 pKa = 9.9ILRR173 pKa = 11.84SGTTADD179 pKa = 4.6MIPLPKK185 pKa = 9.85TFSSINGSNVGMLVSVDD202 pKa = 3.39NVEE205 pKa = 4.66FADD208 pKa = 4.12DD209 pKa = 4.12LAGKK213 pKa = 10.28AYY215 pKa = 10.31FDD217 pKa = 4.6PNEE220 pKa = 4.35SYY222 pKa = 9.17DD223 pKa = 3.76TQRR226 pKa = 11.84TMQACEE232 pKa = 3.86GLSYY236 pKa = 10.6ATFQLEE242 pKa = 4.35TSSFASFKK250 pKa = 10.77QEE252 pKa = 3.77PLPIKK257 pKa = 10.41NGTISAVVVKK267 pKa = 9.62TFDD270 pKa = 3.22GSQVVMALNSINDD283 pKa = 3.45VNFTDD288 pKa = 4.74ARR290 pKa = 11.84CSLLDD295 pKa = 3.44PTDD298 pKa = 3.61FTEE301 pKa = 4.77IFSEE305 pKa = 4.64DD306 pKa = 4.24FEE308 pKa = 4.76TMAANQSINANGWTNYY324 pKa = 9.47IEE326 pKa = 4.55AGNRR330 pKa = 11.84DD331 pKa = 3.12WRR333 pKa = 11.84VVVTTDD339 pKa = 2.86SGNPGSQVASFGAYY353 pKa = 10.09NSGDD357 pKa = 3.51VQNISWLITPGVDD370 pKa = 4.72LDD372 pKa = 3.84ASDD375 pKa = 4.84YY376 pKa = 11.13EE377 pKa = 4.32FLSFEE382 pKa = 4.34TSNSFSDD389 pKa = 4.03GSEE392 pKa = 3.93LEE394 pKa = 4.94LYY396 pKa = 10.28ISTDD400 pKa = 3.1WNGVEE405 pKa = 4.77ADD407 pKa = 3.75VANATWMPLSGTIVSDD423 pKa = 3.35GTFYY427 pKa = 11.06QDD429 pKa = 3.52WISSGSIDD437 pKa = 3.55LSSYY441 pKa = 10.98SGTAHH446 pKa = 6.61VAFKK450 pKa = 11.1YY451 pKa = 10.73VGGGDD456 pKa = 3.34AGIDD460 pKa = 3.43GTYY463 pKa = 10.72EE464 pKa = 4.04LDD466 pKa = 3.62NVSIVANN473 pKa = 3.97
MM1 pKa = 7.17KK2 pKa = 9.86TNKK5 pKa = 9.65FLTLILSFVAVFAMTSCVEE24 pKa = 4.48DD25 pKa = 4.45DD26 pKa = 4.72DD27 pKa = 4.46FTVPSSLGNEE37 pKa = 3.78EE38 pKa = 3.65NAALVEE44 pKa = 4.0LLEE47 pKa = 4.56TSTEE51 pKa = 3.35VDD53 pKa = 2.95MAYY56 pKa = 10.57VQGLYY61 pKa = 10.25TSGTAPEE68 pKa = 4.44QIAADD73 pKa = 4.05IYY75 pKa = 9.63VKK77 pKa = 10.72GYY79 pKa = 10.41VSSSDD84 pKa = 3.2ATGNFYY90 pKa = 11.0KK91 pKa = 10.25EE92 pKa = 5.23FYY94 pKa = 9.55MQDD97 pKa = 3.27TPSNPTKK104 pKa = 10.19ALKK107 pKa = 10.34IIVSQSDD114 pKa = 3.89TYY116 pKa = 11.53NQFNKK121 pKa = 9.74GRR123 pKa = 11.84EE124 pKa = 3.93VYY126 pKa = 10.44INLKK130 pKa = 9.33GLYY133 pKa = 9.51VGEE136 pKa = 4.03EE137 pKa = 3.74RR138 pKa = 11.84VGNGVYY144 pKa = 10.21TIGGSVEE151 pKa = 3.77TDD153 pKa = 3.01QFGTTVQQLDD163 pKa = 3.48EE164 pKa = 4.72HH165 pKa = 6.08QVEE168 pKa = 4.66TKK170 pKa = 9.9ILRR173 pKa = 11.84SGTTADD179 pKa = 4.6MIPLPKK185 pKa = 9.85TFSSINGSNVGMLVSVDD202 pKa = 3.39NVEE205 pKa = 4.66FADD208 pKa = 4.12DD209 pKa = 4.12LAGKK213 pKa = 10.28AYY215 pKa = 10.31FDD217 pKa = 4.6PNEE220 pKa = 4.35SYY222 pKa = 9.17DD223 pKa = 3.76TQRR226 pKa = 11.84TMQACEE232 pKa = 3.86GLSYY236 pKa = 10.6ATFQLEE242 pKa = 4.35TSSFASFKK250 pKa = 10.77QEE252 pKa = 3.77PLPIKK257 pKa = 10.41NGTISAVVVKK267 pKa = 9.62TFDD270 pKa = 3.22GSQVVMALNSINDD283 pKa = 3.45VNFTDD288 pKa = 4.74ARR290 pKa = 11.84CSLLDD295 pKa = 3.44PTDD298 pKa = 3.61FTEE301 pKa = 4.77IFSEE305 pKa = 4.64DD306 pKa = 4.24FEE308 pKa = 4.76TMAANQSINANGWTNYY324 pKa = 9.47IEE326 pKa = 4.55AGNRR330 pKa = 11.84DD331 pKa = 3.12WRR333 pKa = 11.84VVVTTDD339 pKa = 2.86SGNPGSQVASFGAYY353 pKa = 10.09NSGDD357 pKa = 3.51VQNISWLITPGVDD370 pKa = 4.72LDD372 pKa = 3.84ASDD375 pKa = 4.84YY376 pKa = 11.13EE377 pKa = 4.32FLSFEE382 pKa = 4.34TSNSFSDD389 pKa = 4.03GSEE392 pKa = 3.93LEE394 pKa = 4.94LYY396 pKa = 10.28ISTDD400 pKa = 3.1WNGVEE405 pKa = 4.77ADD407 pKa = 3.75VANATWMPLSGTIVSDD423 pKa = 3.35GTFYY427 pKa = 11.06QDD429 pKa = 3.52WISSGSIDD437 pKa = 3.55LSSYY441 pKa = 10.98SGTAHH446 pKa = 6.61VAFKK450 pKa = 11.1YY451 pKa = 10.73VGGGDD456 pKa = 3.34AGIDD460 pKa = 3.43GTYY463 pKa = 10.72EE464 pKa = 4.04LDD466 pKa = 3.62NVSIVANN473 pKa = 3.97
Molecular weight: 51.31 kDa
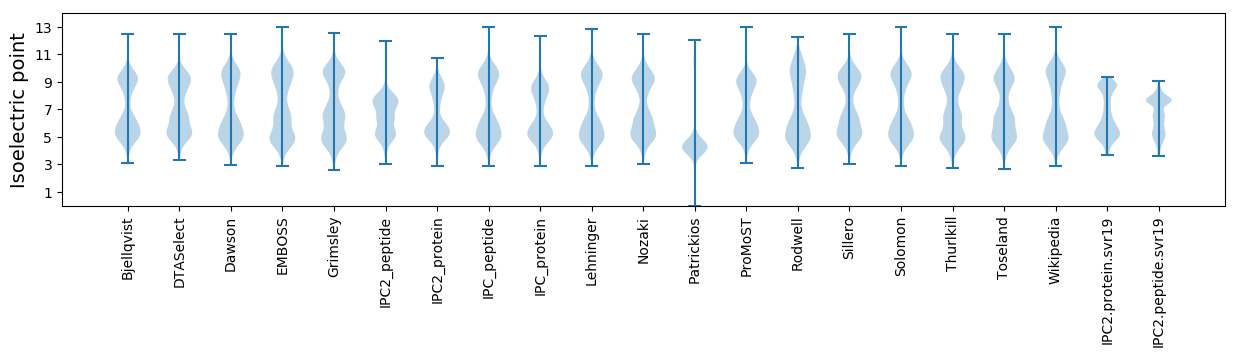
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3YA99|A0A1H3YA99_9FLAO Glyceraldehyde 3-phosphate dehydrogenase OS=Bizionia paragorgiae OX=283786 GN=SAMN04487990_106115 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.92GRR40 pKa = 11.84KK41 pKa = 8.34KK42 pKa = 10.21ISVSSEE48 pKa = 3.74TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.92GRR40 pKa = 11.84KK41 pKa = 8.34KK42 pKa = 10.21ISVSSEE48 pKa = 3.74TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
932437 |
24 |
4616 |
326.1 |
36.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.59 ± 0.043 | 0.764 ± 0.016 |
5.644 ± 0.04 | 6.499 ± 0.047 |
5.139 ± 0.042 | 6.141 ± 0.044 |
1.867 ± 0.025 | 7.789 ± 0.044 |
7.775 ± 0.06 | 9.496 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.08 ± 0.021 | 6.064 ± 0.046 |
3.315 ± 0.026 | 3.414 ± 0.03 |
3.317 ± 0.031 | 6.538 ± 0.039 |
6.212 ± 0.062 | 6.38 ± 0.038 |
0.922 ± 0.014 | 4.053 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
