
Pseudomonas pachastrellae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
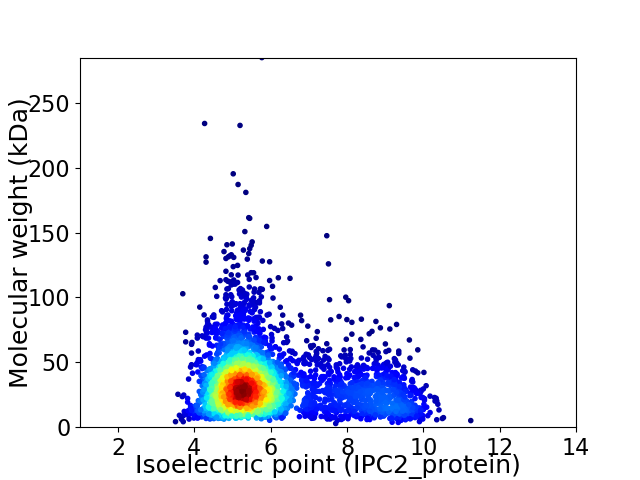
Virtual 2D-PAGE plot for 3588 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S8DDE1|A0A1S8DDE1_9PSED Uncharacterized protein OS=Pseudomonas pachastrellae OX=254161 GN=BXT89_16915 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 10.34RR3 pKa = 11.84FSALLLSSMLLSTAAQAQDD22 pKa = 3.3SFKK25 pKa = 10.87VCWSIYY31 pKa = 9.34AGWMPWAYY39 pKa = 9.76GAEE42 pKa = 4.02SGIVKK47 pKa = 9.99KK48 pKa = 9.67WADD51 pKa = 3.62KK52 pKa = 10.9YY53 pKa = 11.22DD54 pKa = 3.54INIDD58 pKa = 3.49VVQINDD64 pKa = 3.86YY65 pKa = 10.75IEE67 pKa = 5.44SINQYY72 pKa = 8.45TVGEE76 pKa = 4.29FDD78 pKa = 3.7ACTMTNMDD86 pKa = 4.98ALTIPAAGGVDD97 pKa = 3.65STALIVGDD105 pKa = 4.41FSNGNDD111 pKa = 4.1GIVLKK116 pKa = 11.12GEE118 pKa = 4.27SEE120 pKa = 3.89LAAIKK125 pKa = 9.67GQNVNLVEE133 pKa = 4.59LSVSHH138 pKa = 6.06YY139 pKa = 11.02LLARR143 pKa = 11.84ALDD146 pKa = 3.95SVGLSEE152 pKa = 6.34ADD154 pKa = 3.43VQVVNTSDD162 pKa = 3.46ADD164 pKa = 3.64MVAVYY169 pKa = 8.45GTDD172 pKa = 3.32DD173 pKa = 3.72VTSVATWNPLLSEE186 pKa = 4.75ISQQPSSTVVFNSAQIPGEE205 pKa = 4.27IIDD208 pKa = 4.62LLVINTDD215 pKa = 3.17TLAAHH220 pKa = 6.98PEE222 pKa = 3.97LGKK225 pKa = 10.71ALTGAWYY232 pKa = 9.7EE233 pKa = 4.07IMATMSIDD241 pKa = 3.61SEE243 pKa = 4.39QGVAARR249 pKa = 11.84SYY251 pKa = 8.95MAEE254 pKa = 3.85ASGTDD259 pKa = 3.47LAGYY263 pKa = 7.75EE264 pKa = 4.1AQLAATEE271 pKa = 4.24MFYY274 pKa = 10.76TPAAALEE281 pKa = 4.41LTNSPQLKK289 pKa = 8.2QTMQYY294 pKa = 9.82VAEE297 pKa = 4.38FSFDD301 pKa = 3.28HH302 pKa = 6.69GLLGDD307 pKa = 4.51GAPDD311 pKa = 3.22AGFIGVQMPAGDD323 pKa = 4.18YY324 pKa = 11.17GDD326 pKa = 3.99SSNLKK331 pKa = 10.29LRR333 pKa = 11.84FDD335 pKa = 3.76PTYY338 pKa = 10.37MQMAADD344 pKa = 3.83NQLL347 pKa = 3.03
MM1 pKa = 7.35KK2 pKa = 10.34RR3 pKa = 11.84FSALLLSSMLLSTAAQAQDD22 pKa = 3.3SFKK25 pKa = 10.87VCWSIYY31 pKa = 9.34AGWMPWAYY39 pKa = 9.76GAEE42 pKa = 4.02SGIVKK47 pKa = 9.99KK48 pKa = 9.67WADD51 pKa = 3.62KK52 pKa = 10.9YY53 pKa = 11.22DD54 pKa = 3.54INIDD58 pKa = 3.49VVQINDD64 pKa = 3.86YY65 pKa = 10.75IEE67 pKa = 5.44SINQYY72 pKa = 8.45TVGEE76 pKa = 4.29FDD78 pKa = 3.7ACTMTNMDD86 pKa = 4.98ALTIPAAGGVDD97 pKa = 3.65STALIVGDD105 pKa = 4.41FSNGNDD111 pKa = 4.1GIVLKK116 pKa = 11.12GEE118 pKa = 4.27SEE120 pKa = 3.89LAAIKK125 pKa = 9.67GQNVNLVEE133 pKa = 4.59LSVSHH138 pKa = 6.06YY139 pKa = 11.02LLARR143 pKa = 11.84ALDD146 pKa = 3.95SVGLSEE152 pKa = 6.34ADD154 pKa = 3.43VQVVNTSDD162 pKa = 3.46ADD164 pKa = 3.64MVAVYY169 pKa = 8.45GTDD172 pKa = 3.32DD173 pKa = 3.72VTSVATWNPLLSEE186 pKa = 4.75ISQQPSSTVVFNSAQIPGEE205 pKa = 4.27IIDD208 pKa = 4.62LLVINTDD215 pKa = 3.17TLAAHH220 pKa = 6.98PEE222 pKa = 3.97LGKK225 pKa = 10.71ALTGAWYY232 pKa = 9.7EE233 pKa = 4.07IMATMSIDD241 pKa = 3.61SEE243 pKa = 4.39QGVAARR249 pKa = 11.84SYY251 pKa = 8.95MAEE254 pKa = 3.85ASGTDD259 pKa = 3.47LAGYY263 pKa = 7.75EE264 pKa = 4.1AQLAATEE271 pKa = 4.24MFYY274 pKa = 10.76TPAAALEE281 pKa = 4.41LTNSPQLKK289 pKa = 8.2QTMQYY294 pKa = 9.82VAEE297 pKa = 4.38FSFDD301 pKa = 3.28HH302 pKa = 6.69GLLGDD307 pKa = 4.51GAPDD311 pKa = 3.22AGFIGVQMPAGDD323 pKa = 4.18YY324 pKa = 11.17GDD326 pKa = 3.99SSNLKK331 pKa = 10.29LRR333 pKa = 11.84FDD335 pKa = 3.76PTYY338 pKa = 10.37MQMAADD344 pKa = 3.83NQLL347 pKa = 3.03
Molecular weight: 37.11 kDa
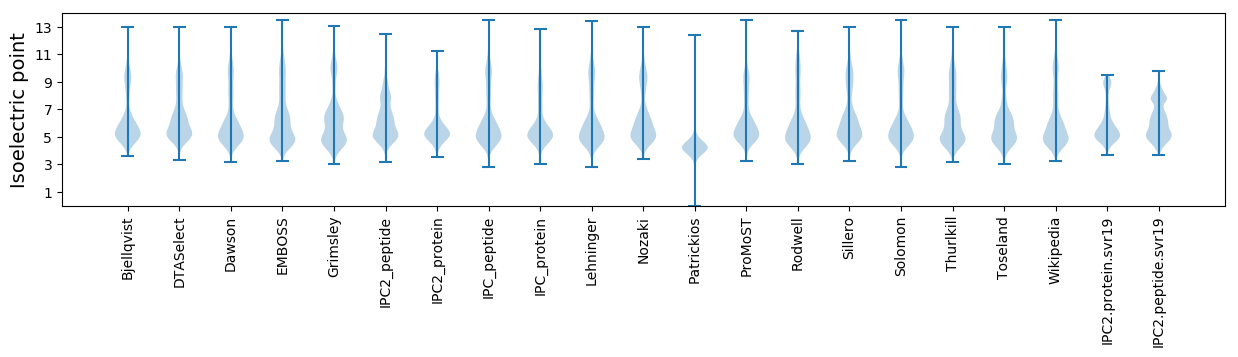
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S8DE03|A0A1S8DE03_9PSED Uncharacterized protein OS=Pseudomonas pachastrellae OX=254161 GN=BXT89_16315 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1158881 |
23 |
2623 |
323.0 |
35.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.06 ± 0.053 | 1.028 ± 0.013 |
5.679 ± 0.031 | 6.046 ± 0.04 |
3.442 ± 0.022 | 7.819 ± 0.037 |
2.227 ± 0.02 | 4.691 ± 0.032 |
2.965 ± 0.04 | 11.827 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.423 ± 0.02 | 2.953 ± 0.022 |
4.761 ± 0.03 | 4.962 ± 0.04 |
6.684 ± 0.034 | 5.783 ± 0.028 |
4.718 ± 0.026 | 6.973 ± 0.035 |
1.441 ± 0.017 | 2.518 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
