
Nocardioides sp. AX2bis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
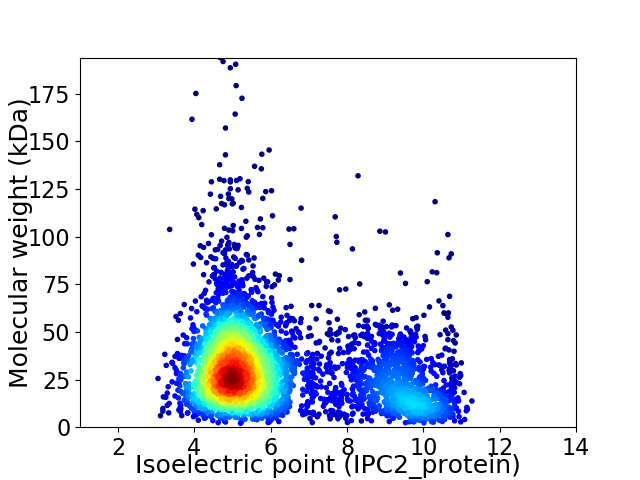
Virtual 2D-PAGE plot for 4362 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A653VG68|A0A653VG68_9ACTN Uncharacterized protein OS=Nocardioides sp. AX2bis OX=2653157 GN=NOCARDAX2BIS_400155 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 10.43NPEE5 pKa = 3.81RR6 pKa = 11.84RR7 pKa = 11.84GTMKK11 pKa = 9.95TRR13 pKa = 11.84RR14 pKa = 11.84LLATAAVATLALTACSSKK32 pKa = 9.98STDD35 pKa = 3.41SAEE38 pKa = 4.36GGAAAPGVTDD48 pKa = 3.64DD49 pKa = 4.52TISLGVLTDD58 pKa = 3.15QSGPFIQLGTGTVDD72 pKa = 3.78GHH74 pKa = 6.09QIWADD79 pKa = 3.58EE80 pKa = 4.14VNAAGGICGRR90 pKa = 11.84QIEE93 pKa = 4.85LVVRR97 pKa = 11.84DD98 pKa = 4.07HH99 pKa = 7.34GYY101 pKa = 9.67QADD104 pKa = 3.96AGVLAYY110 pKa = 10.29QEE112 pKa = 4.76LLPEE116 pKa = 4.32VLGFMQILGSPVIAALDD133 pKa = 3.38QQLIDD138 pKa = 3.97NEE140 pKa = 4.59TTSVALSWSSFLLKK154 pKa = 10.5NPYY157 pKa = 9.71VVIPGTTYY165 pKa = 10.79DD166 pKa = 3.39VEE168 pKa = 4.55MVNGLSYY175 pKa = 11.11LLEE178 pKa = 4.13QGEE181 pKa = 4.31IAEE184 pKa = 4.49GDD186 pKa = 3.78TVGFVYY192 pKa = 10.92LEE194 pKa = 4.11GEE196 pKa = 4.09YY197 pKa = 11.26GEE199 pKa = 4.95NGLLGAEE206 pKa = 4.45YY207 pKa = 9.71FAEE210 pKa = 4.0QHH212 pKa = 6.97DD213 pKa = 4.12ITLEE217 pKa = 3.98AVKK220 pKa = 10.39VLPTDD225 pKa = 3.22TDD227 pKa = 3.32LRR229 pKa = 11.84NVVTGLRR236 pKa = 11.84GADD239 pKa = 3.3VSAIGLTTTPSQTLSVATVAAQLGLDD265 pKa = 3.68VPLVGNNPTFAPAILTEE282 pKa = 4.23DD283 pKa = 3.14TAAALEE289 pKa = 3.96NYY291 pKa = 9.45YY292 pKa = 10.73LVGSATPYY300 pKa = 10.72SSDD303 pKa = 3.35VPKK306 pKa = 10.56AAEE309 pKa = 3.71VATAYY314 pKa = 10.27EE315 pKa = 4.33EE316 pKa = 4.34LGKK319 pKa = 10.17SEE321 pKa = 4.45PANAGIPYY329 pKa = 9.42GYY331 pKa = 10.41AIGEE335 pKa = 3.72IWGQVLEE342 pKa = 4.65ASCDD346 pKa = 3.53DD347 pKa = 3.75LTRR350 pKa = 11.84SGIQDD355 pKa = 3.71ALASSTDD362 pKa = 3.23VTTDD366 pKa = 3.29EE367 pKa = 5.04LVADD371 pKa = 5.35LDD373 pKa = 4.11LTSPGSPATRR383 pKa = 11.84EE384 pKa = 4.17VYY386 pKa = 10.1IATADD391 pKa = 3.26IDD393 pKa = 4.19AEE395 pKa = 4.57GGLTQQGDD403 pKa = 4.32LFTSPDD409 pKa = 3.49AEE411 pKa = 4.17SYY413 pKa = 9.88VAPEE417 pKa = 4.06EE418 pKa = 4.15QQ419 pKa = 3.43
MM1 pKa = 7.66KK2 pKa = 10.43NPEE5 pKa = 3.81RR6 pKa = 11.84RR7 pKa = 11.84GTMKK11 pKa = 9.95TRR13 pKa = 11.84RR14 pKa = 11.84LLATAAVATLALTACSSKK32 pKa = 9.98STDD35 pKa = 3.41SAEE38 pKa = 4.36GGAAAPGVTDD48 pKa = 3.64DD49 pKa = 4.52TISLGVLTDD58 pKa = 3.15QSGPFIQLGTGTVDD72 pKa = 3.78GHH74 pKa = 6.09QIWADD79 pKa = 3.58EE80 pKa = 4.14VNAAGGICGRR90 pKa = 11.84QIEE93 pKa = 4.85LVVRR97 pKa = 11.84DD98 pKa = 4.07HH99 pKa = 7.34GYY101 pKa = 9.67QADD104 pKa = 3.96AGVLAYY110 pKa = 10.29QEE112 pKa = 4.76LLPEE116 pKa = 4.32VLGFMQILGSPVIAALDD133 pKa = 3.38QQLIDD138 pKa = 3.97NEE140 pKa = 4.59TTSVALSWSSFLLKK154 pKa = 10.5NPYY157 pKa = 9.71VVIPGTTYY165 pKa = 10.79DD166 pKa = 3.39VEE168 pKa = 4.55MVNGLSYY175 pKa = 11.11LLEE178 pKa = 4.13QGEE181 pKa = 4.31IAEE184 pKa = 4.49GDD186 pKa = 3.78TVGFVYY192 pKa = 10.92LEE194 pKa = 4.11GEE196 pKa = 4.09YY197 pKa = 11.26GEE199 pKa = 4.95NGLLGAEE206 pKa = 4.45YY207 pKa = 9.71FAEE210 pKa = 4.0QHH212 pKa = 6.97DD213 pKa = 4.12ITLEE217 pKa = 3.98AVKK220 pKa = 10.39VLPTDD225 pKa = 3.22TDD227 pKa = 3.32LRR229 pKa = 11.84NVVTGLRR236 pKa = 11.84GADD239 pKa = 3.3VSAIGLTTTPSQTLSVATVAAQLGLDD265 pKa = 3.68VPLVGNNPTFAPAILTEE282 pKa = 4.23DD283 pKa = 3.14TAAALEE289 pKa = 3.96NYY291 pKa = 9.45YY292 pKa = 10.73LVGSATPYY300 pKa = 10.72SSDD303 pKa = 3.35VPKK306 pKa = 10.56AAEE309 pKa = 3.71VATAYY314 pKa = 10.27EE315 pKa = 4.33EE316 pKa = 4.34LGKK319 pKa = 10.17SEE321 pKa = 4.45PANAGIPYY329 pKa = 9.42GYY331 pKa = 10.41AIGEE335 pKa = 3.72IWGQVLEE342 pKa = 4.65ASCDD346 pKa = 3.53DD347 pKa = 3.75LTRR350 pKa = 11.84SGIQDD355 pKa = 3.71ALASSTDD362 pKa = 3.23VTTDD366 pKa = 3.29EE367 pKa = 5.04LVADD371 pKa = 5.35LDD373 pKa = 4.11LTSPGSPATRR383 pKa = 11.84EE384 pKa = 4.17VYY386 pKa = 10.1IATADD391 pKa = 3.26IDD393 pKa = 4.19AEE395 pKa = 4.57GGLTQQGDD403 pKa = 4.32LFTSPDD409 pKa = 3.49AEE411 pKa = 4.17SYY413 pKa = 9.88VAPEE417 pKa = 4.06EE418 pKa = 4.15QQ419 pKa = 3.43
Molecular weight: 43.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A653Q5E6|A0A653Q5E6_9ACTN Dipeptidyl aminopeptidase BI OS=Nocardioides sp. AX2bis OX=2653157 GN=dapb PE=4 SV=1
MM1 pKa = 7.43ARR3 pKa = 11.84GLRR6 pKa = 11.84RR7 pKa = 11.84PARR10 pKa = 11.84RR11 pKa = 11.84VGAAVRR17 pKa = 11.84HH18 pKa = 5.92RR19 pKa = 11.84SRR21 pKa = 11.84ALGGPRR27 pKa = 11.84QAAGRR32 pKa = 11.84GRR34 pKa = 11.84QGRR37 pKa = 11.84GRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84GHH44 pKa = 5.98VLLLRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84QHH53 pKa = 6.79RR54 pKa = 11.84LGLHH58 pKa = 5.62AAGVLGRR65 pKa = 11.84HH66 pKa = 5.4GEE68 pKa = 3.91HH69 pKa = 6.72RR70 pKa = 11.84RR71 pKa = 11.84LAGLRR76 pKa = 11.84RR77 pKa = 11.84GPPGPARR84 pKa = 11.84EE85 pKa = 3.8AHH87 pKa = 5.47RR88 pKa = 11.84RR89 pKa = 11.84GRR91 pKa = 11.84LTRR94 pKa = 11.84TQHH97 pKa = 5.54PRR99 pKa = 11.84RR100 pKa = 11.84PVATSVAAGRR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84SRR114 pKa = 11.84TGRR117 pKa = 11.84PGPGAPVHH125 pKa = 6.19
MM1 pKa = 7.43ARR3 pKa = 11.84GLRR6 pKa = 11.84RR7 pKa = 11.84PARR10 pKa = 11.84RR11 pKa = 11.84VGAAVRR17 pKa = 11.84HH18 pKa = 5.92RR19 pKa = 11.84SRR21 pKa = 11.84ALGGPRR27 pKa = 11.84QAAGRR32 pKa = 11.84GRR34 pKa = 11.84QGRR37 pKa = 11.84GRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84GHH44 pKa = 5.98VLLLRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84QHH53 pKa = 6.79RR54 pKa = 11.84LGLHH58 pKa = 5.62AAGVLGRR65 pKa = 11.84HH66 pKa = 5.4GEE68 pKa = 3.91HH69 pKa = 6.72RR70 pKa = 11.84RR71 pKa = 11.84LAGLRR76 pKa = 11.84RR77 pKa = 11.84GPPGPARR84 pKa = 11.84EE85 pKa = 3.8AHH87 pKa = 5.47RR88 pKa = 11.84RR89 pKa = 11.84GRR91 pKa = 11.84LTRR94 pKa = 11.84TQHH97 pKa = 5.54PRR99 pKa = 11.84RR100 pKa = 11.84PVATSVAAGRR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84SRR114 pKa = 11.84TGRR117 pKa = 11.84PGPGAPVHH125 pKa = 6.19
Molecular weight: 13.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1328517 |
20 |
1851 |
304.6 |
32.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.489 ± 0.051 | 0.73 ± 0.011 |
6.686 ± 0.038 | 5.594 ± 0.033 |
2.423 ± 0.026 | 9.813 ± 0.041 |
2.252 ± 0.022 | 2.539 ± 0.029 |
1.479 ± 0.025 | 10.353 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.685 ± 0.016 | 1.446 ± 0.018 |
6.134 ± 0.038 | 2.663 ± 0.018 |
8.59 ± 0.065 | 4.925 ± 0.031 |
6.097 ± 0.032 | 9.888 ± 0.044 |
1.468 ± 0.018 | 1.747 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
