
Macropodid alphaherpesvirus 1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Simplexvirus
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 71 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0Y0AC57|A0A0Y0AC57_9ALPH Tegument protein OS=Macropodid alphaherpesvirus 1 OX=137443 GN=UL14 PE=3 SV=1
MM1 pKa = 7.65SPRR4 pKa = 11.84CSVRR8 pKa = 11.84PHH10 pKa = 6.48EE11 pKa = 4.51SVEE14 pKa = 3.98ALLEE18 pKa = 4.38PGAVQEE24 pKa = 4.3TSDD27 pKa = 4.05AEE29 pKa = 4.29EE30 pKa = 3.95VDD32 pKa = 3.44SDD34 pKa = 4.06EE35 pKa = 4.46EE36 pKa = 3.96QSEE39 pKa = 4.4GGEE42 pKa = 4.15SSEE45 pKa = 4.18EE46 pKa = 3.92DD47 pKa = 3.16SSEE50 pKa = 4.11EE51 pKa = 3.75NSEE54 pKa = 4.11LSDD57 pKa = 3.65EE58 pKa = 4.28EE59 pKa = 4.4FSDD62 pKa = 4.6GEE64 pKa = 4.16EE65 pKa = 4.89SSDD68 pKa = 3.3EE69 pKa = 3.87DD70 pKa = 3.74DD71 pKa = 3.43RR72 pKa = 11.84RR73 pKa = 11.84GRR75 pKa = 11.84TRR77 pKa = 11.84RR78 pKa = 11.84QSTKK82 pKa = 9.07TAVLEE87 pKa = 4.1FLDD90 pKa = 4.46LEE92 pKa = 4.71AIEE95 pKa = 4.61VGGGSPEE102 pKa = 4.12YY103 pKa = 10.83GCEE106 pKa = 3.52DD107 pKa = 3.94DD108 pKa = 5.41YY109 pKa = 11.68YY110 pKa = 11.55DD111 pKa = 3.86RR112 pKa = 11.84YY113 pKa = 10.89EE114 pKa = 4.12VFSEE118 pKa = 3.97EE119 pKa = 4.66DD120 pKa = 3.03SDD122 pKa = 4.07EE123 pKa = 4.46GVSEE127 pKa = 4.36EE128 pKa = 4.25DD129 pKa = 3.48HH130 pKa = 7.18KK131 pKa = 11.56DD132 pKa = 3.39ALGCGPSDD140 pKa = 4.15SKK142 pKa = 11.64LPDD145 pKa = 3.82LLSDD149 pKa = 4.13VPEE152 pKa = 4.5HH153 pKa = 6.68PANKK157 pKa = 10.19GEE159 pKa = 4.15SRR161 pKa = 11.84SNRR164 pKa = 11.84EE165 pKa = 3.19ISEE168 pKa = 4.38DD169 pKa = 3.45EE170 pKa = 4.6SICEE174 pKa = 3.76EE175 pKa = 4.08LVGFRR180 pKa = 11.84HH181 pKa = 6.4KK182 pKa = 10.27RR183 pKa = 11.84KK184 pKa = 8.52KK185 pKa = 7.56WHH187 pKa = 5.51VVEE190 pKa = 5.1SSSEE194 pKa = 3.93SDD196 pKa = 3.3EE197 pKa = 5.94DD198 pKa = 3.9SDD200 pKa = 4.88EE201 pKa = 4.65PEE203 pKa = 4.32STSKK207 pKa = 10.12PRR209 pKa = 11.84KK210 pKa = 8.03LLKK213 pKa = 10.05RR214 pKa = 11.84LRR216 pKa = 11.84RR217 pKa = 11.84QVDD220 pKa = 3.59GEE222 pKa = 4.5GNIASQRR229 pKa = 11.84SATSEE234 pKa = 3.46GRR236 pKa = 11.84QIVSGPDD243 pKa = 3.39NQPGEE248 pKa = 4.36PGASGRR254 pKa = 11.84RR255 pKa = 11.84PVKK258 pKa = 10.56RR259 pKa = 11.84NPTWSEE265 pKa = 3.47LQQNINQLFRR275 pKa = 11.84FLRR278 pKa = 11.84DD279 pKa = 3.26KK280 pKa = 11.05PNGRR284 pKa = 11.84GRR286 pKa = 11.84IDD288 pKa = 3.38RR289 pKa = 11.84LRR291 pKa = 11.84DD292 pKa = 3.57VVIDD296 pKa = 4.14IYY298 pKa = 11.62LMGYY302 pKa = 7.11EE303 pKa = 4.55RR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84LDD308 pKa = 3.22PTGWGVLLQTTGHH321 pKa = 5.98RR322 pKa = 11.84TDD324 pKa = 3.93AVRR327 pKa = 11.84DD328 pKa = 4.17DD329 pKa = 3.72IGGVEE334 pKa = 4.69CLVGVPGPTDD344 pKa = 3.26SLRR347 pKa = 11.84PLAGKK352 pKa = 10.32SYY354 pKa = 8.92GQCGSGGVPEE364 pKa = 4.62TSDD367 pKa = 3.16IDD369 pKa = 3.94SDD371 pKa = 3.71ASTVVVNTKK380 pKa = 10.04GRR382 pKa = 11.84WKK384 pKa = 9.66TSGNAARR391 pKa = 11.84LDD393 pKa = 3.54EE394 pKa = 4.33EE395 pKa = 4.67TRR397 pKa = 11.84VGSPLRR403 pKa = 11.84DD404 pKa = 3.27SPRR407 pKa = 11.84RR408 pKa = 11.84GEE410 pKa = 4.37LDD412 pKa = 3.43ASPSMATQPWPSMVVADD429 pKa = 4.72TDD431 pKa = 4.27STDD434 pKa = 3.03LWSQSTGQTFPVFKK448 pKa = 9.53DD449 pKa = 3.3TKK451 pKa = 9.37GCCEE455 pKa = 3.84IQFPAPRR462 pKa = 11.84RR463 pKa = 3.7
MM1 pKa = 7.65SPRR4 pKa = 11.84CSVRR8 pKa = 11.84PHH10 pKa = 6.48EE11 pKa = 4.51SVEE14 pKa = 3.98ALLEE18 pKa = 4.38PGAVQEE24 pKa = 4.3TSDD27 pKa = 4.05AEE29 pKa = 4.29EE30 pKa = 3.95VDD32 pKa = 3.44SDD34 pKa = 4.06EE35 pKa = 4.46EE36 pKa = 3.96QSEE39 pKa = 4.4GGEE42 pKa = 4.15SSEE45 pKa = 4.18EE46 pKa = 3.92DD47 pKa = 3.16SSEE50 pKa = 4.11EE51 pKa = 3.75NSEE54 pKa = 4.11LSDD57 pKa = 3.65EE58 pKa = 4.28EE59 pKa = 4.4FSDD62 pKa = 4.6GEE64 pKa = 4.16EE65 pKa = 4.89SSDD68 pKa = 3.3EE69 pKa = 3.87DD70 pKa = 3.74DD71 pKa = 3.43RR72 pKa = 11.84RR73 pKa = 11.84GRR75 pKa = 11.84TRR77 pKa = 11.84RR78 pKa = 11.84QSTKK82 pKa = 9.07TAVLEE87 pKa = 4.1FLDD90 pKa = 4.46LEE92 pKa = 4.71AIEE95 pKa = 4.61VGGGSPEE102 pKa = 4.12YY103 pKa = 10.83GCEE106 pKa = 3.52DD107 pKa = 3.94DD108 pKa = 5.41YY109 pKa = 11.68YY110 pKa = 11.55DD111 pKa = 3.86RR112 pKa = 11.84YY113 pKa = 10.89EE114 pKa = 4.12VFSEE118 pKa = 3.97EE119 pKa = 4.66DD120 pKa = 3.03SDD122 pKa = 4.07EE123 pKa = 4.46GVSEE127 pKa = 4.36EE128 pKa = 4.25DD129 pKa = 3.48HH130 pKa = 7.18KK131 pKa = 11.56DD132 pKa = 3.39ALGCGPSDD140 pKa = 4.15SKK142 pKa = 11.64LPDD145 pKa = 3.82LLSDD149 pKa = 4.13VPEE152 pKa = 4.5HH153 pKa = 6.68PANKK157 pKa = 10.19GEE159 pKa = 4.15SRR161 pKa = 11.84SNRR164 pKa = 11.84EE165 pKa = 3.19ISEE168 pKa = 4.38DD169 pKa = 3.45EE170 pKa = 4.6SICEE174 pKa = 3.76EE175 pKa = 4.08LVGFRR180 pKa = 11.84HH181 pKa = 6.4KK182 pKa = 10.27RR183 pKa = 11.84KK184 pKa = 8.52KK185 pKa = 7.56WHH187 pKa = 5.51VVEE190 pKa = 5.1SSSEE194 pKa = 3.93SDD196 pKa = 3.3EE197 pKa = 5.94DD198 pKa = 3.9SDD200 pKa = 4.88EE201 pKa = 4.65PEE203 pKa = 4.32STSKK207 pKa = 10.12PRR209 pKa = 11.84KK210 pKa = 8.03LLKK213 pKa = 10.05RR214 pKa = 11.84LRR216 pKa = 11.84RR217 pKa = 11.84QVDD220 pKa = 3.59GEE222 pKa = 4.5GNIASQRR229 pKa = 11.84SATSEE234 pKa = 3.46GRR236 pKa = 11.84QIVSGPDD243 pKa = 3.39NQPGEE248 pKa = 4.36PGASGRR254 pKa = 11.84RR255 pKa = 11.84PVKK258 pKa = 10.56RR259 pKa = 11.84NPTWSEE265 pKa = 3.47LQQNINQLFRR275 pKa = 11.84FLRR278 pKa = 11.84DD279 pKa = 3.26KK280 pKa = 11.05PNGRR284 pKa = 11.84GRR286 pKa = 11.84IDD288 pKa = 3.38RR289 pKa = 11.84LRR291 pKa = 11.84DD292 pKa = 3.57VVIDD296 pKa = 4.14IYY298 pKa = 11.62LMGYY302 pKa = 7.11EE303 pKa = 4.55RR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84LDD308 pKa = 3.22PTGWGVLLQTTGHH321 pKa = 5.98RR322 pKa = 11.84TDD324 pKa = 3.93AVRR327 pKa = 11.84DD328 pKa = 4.17DD329 pKa = 3.72IGGVEE334 pKa = 4.69CLVGVPGPTDD344 pKa = 3.26SLRR347 pKa = 11.84PLAGKK352 pKa = 10.32SYY354 pKa = 8.92GQCGSGGVPEE364 pKa = 4.62TSDD367 pKa = 3.16IDD369 pKa = 3.94SDD371 pKa = 3.71ASTVVVNTKK380 pKa = 10.04GRR382 pKa = 11.84WKK384 pKa = 9.66TSGNAARR391 pKa = 11.84LDD393 pKa = 3.54EE394 pKa = 4.33EE395 pKa = 4.67TRR397 pKa = 11.84VGSPLRR403 pKa = 11.84DD404 pKa = 3.27SPRR407 pKa = 11.84RR408 pKa = 11.84GEE410 pKa = 4.37LDD412 pKa = 3.43ASPSMATQPWPSMVVADD429 pKa = 4.72TDD431 pKa = 4.27STDD434 pKa = 3.03LWSQSTGQTFPVFKK448 pKa = 9.53DD449 pKa = 3.3TKK451 pKa = 9.37GCCEE455 pKa = 3.84IQFPAPRR462 pKa = 11.84RR463 pKa = 3.7
Molecular weight: 51.11 kDa
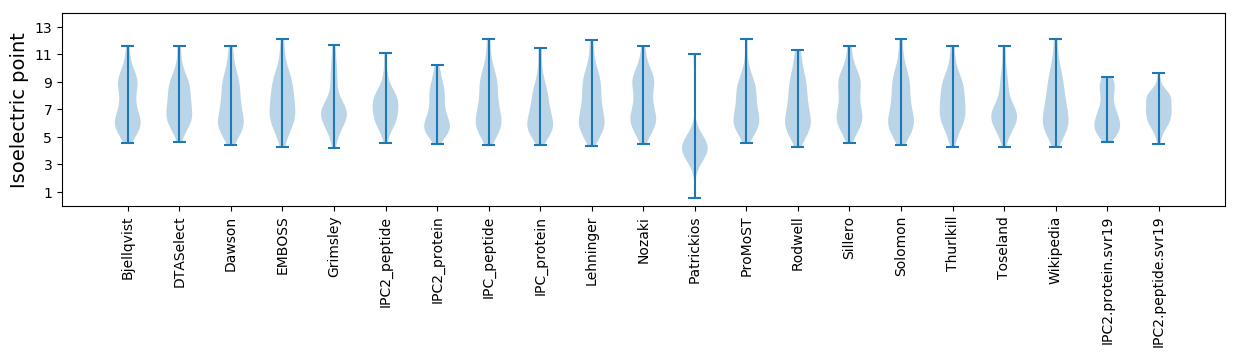
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A120HUJ2|A0A120HUJ2_9ALPH Isoform of A0A0Y0A7L1 GG OS=Macropodid alphaherpesvirus 1 OX=137443 GN=US4 PE=4 SV=1
MM1 pKa = 7.85PATSPSIPLPRR12 pKa = 11.84ATPIRR17 pKa = 11.84IRR19 pKa = 11.84TRR21 pKa = 11.84PRR23 pKa = 11.84RR24 pKa = 11.84PPIRR28 pKa = 11.84PTGLTNARR36 pKa = 11.84RR37 pKa = 11.84SPGPGTGDD45 pKa = 3.0LSLRR49 pKa = 11.84SRR51 pKa = 11.84VVRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 7.98RR57 pKa = 11.84TCVRR61 pKa = 11.84SAPTPFTDD69 pKa = 4.1AVRR72 pKa = 11.84ATTFPCLHH80 pKa = 6.3NFCVACLKK88 pKa = 9.67TWLPQHH94 pKa = 5.82NRR96 pKa = 11.84CPLCNLSVSYY106 pKa = 10.28LIVDD110 pKa = 3.69VRR112 pKa = 11.84PDD114 pKa = 3.44GTFSTIPVVRR124 pKa = 11.84QQDD127 pKa = 4.18LYY129 pKa = 11.11GEE131 pKa = 4.06PAAAVRR137 pKa = 11.84AGTAVDD143 pKa = 5.06FIWTGAARR151 pKa = 11.84QAPASVSAGGYY162 pKa = 4.93TVRR165 pKa = 11.84ARR167 pKa = 11.84SPRR170 pKa = 11.84RR171 pKa = 11.84SIGSSDD177 pKa = 3.25GANRR181 pKa = 11.84PTRR184 pKa = 11.84PVGRR188 pKa = 11.84PRR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84RR194 pKa = 11.84SNQVSPRR201 pKa = 11.84ADD203 pKa = 4.05PIVIDD208 pKa = 5.13DD209 pKa = 5.19DD210 pKa = 4.58EE211 pKa = 7.53DD212 pKa = 5.48DD213 pKa = 4.73GAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPRR259 pKa = 11.84PHH261 pKa = 6.5ATVTPQPRR269 pKa = 11.84TLWTSEE275 pKa = 3.77TGTSGLEE282 pKa = 3.99GPRR285 pKa = 11.84ATRR288 pKa = 11.84VRR290 pKa = 11.84VEE292 pKa = 3.99VLCNIARR299 pKa = 11.84TEE301 pKa = 4.1EE302 pKa = 4.12PQRR305 pKa = 11.84TPTASPDD312 pKa = 3.26ATPVHH317 pKa = 6.74SSTVPEE323 pKa = 4.1EE324 pKa = 4.36TPAGASSVPAPAPVRR339 pKa = 11.84KK340 pKa = 9.46RR341 pKa = 11.84RR342 pKa = 11.84LATEE346 pKa = 4.1NRR348 pKa = 11.84ASSPDD353 pKa = 3.37TRR355 pKa = 11.84GRR357 pKa = 11.84PAADD361 pKa = 3.56PRR363 pKa = 11.84PGTSGAAEE371 pKa = 4.06EE372 pKa = 4.97GEE374 pKa = 4.41EE375 pKa = 4.25EE376 pKa = 4.72DD377 pKa = 5.63GEE379 pKa = 4.93GPTPSDD385 pKa = 3.16PRR387 pKa = 11.84AGRR390 pKa = 11.84DD391 pKa = 3.43QPCRR395 pKa = 11.84KK396 pKa = 9.3CVRR399 pKa = 11.84KK400 pKa = 7.34THH402 pKa = 6.03HH403 pKa = 6.75RR404 pKa = 11.84SALAAEE410 pKa = 4.5RR411 pKa = 11.84PVEE414 pKa = 4.33GSFLQYY420 pKa = 10.45RR421 pKa = 11.84PLVGLSSAVAMAPFLNKK438 pKa = 9.33TVTGDD443 pKa = 3.52CLPIVDD449 pKa = 5.23LEE451 pKa = 4.3SGEE454 pKa = 3.81IGAYY458 pKa = 9.55VVLTHH463 pKa = 6.12QDD465 pKa = 3.2CNMARR470 pKa = 11.84ALAEE474 pKa = 4.36ARR476 pKa = 11.84STWLDD481 pKa = 3.61DD482 pKa = 3.48TSLPLTNPRR491 pKa = 11.84GATGSDD497 pKa = 3.57VPRR500 pKa = 11.84EE501 pKa = 4.11RR502 pKa = 11.84PGSPFRR508 pKa = 11.84LWSTSRR514 pKa = 11.84GTLLFRR520 pKa = 11.84RR521 pKa = 11.84DD522 pKa = 2.94GTLYY526 pKa = 10.65GRR528 pKa = 11.84QSPFGGVQLHH538 pKa = 6.55PWDD541 pKa = 3.83PP542 pKa = 3.55
MM1 pKa = 7.85PATSPSIPLPRR12 pKa = 11.84ATPIRR17 pKa = 11.84IRR19 pKa = 11.84TRR21 pKa = 11.84PRR23 pKa = 11.84RR24 pKa = 11.84PPIRR28 pKa = 11.84PTGLTNARR36 pKa = 11.84RR37 pKa = 11.84SPGPGTGDD45 pKa = 3.0LSLRR49 pKa = 11.84SRR51 pKa = 11.84VVRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 7.98RR57 pKa = 11.84TCVRR61 pKa = 11.84SAPTPFTDD69 pKa = 4.1AVRR72 pKa = 11.84ATTFPCLHH80 pKa = 6.3NFCVACLKK88 pKa = 9.67TWLPQHH94 pKa = 5.82NRR96 pKa = 11.84CPLCNLSVSYY106 pKa = 10.28LIVDD110 pKa = 3.69VRR112 pKa = 11.84PDD114 pKa = 3.44GTFSTIPVVRR124 pKa = 11.84QQDD127 pKa = 4.18LYY129 pKa = 11.11GEE131 pKa = 4.06PAAAVRR137 pKa = 11.84AGTAVDD143 pKa = 5.06FIWTGAARR151 pKa = 11.84QAPASVSAGGYY162 pKa = 4.93TVRR165 pKa = 11.84ARR167 pKa = 11.84SPRR170 pKa = 11.84RR171 pKa = 11.84SIGSSDD177 pKa = 3.25GANRR181 pKa = 11.84PTRR184 pKa = 11.84PVGRR188 pKa = 11.84PRR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84RR194 pKa = 11.84SNQVSPRR201 pKa = 11.84ADD203 pKa = 4.05PIVIDD208 pKa = 5.13DD209 pKa = 5.19DD210 pKa = 4.58EE211 pKa = 7.53DD212 pKa = 5.48DD213 pKa = 4.73GAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPAPRR259 pKa = 11.84PHH261 pKa = 6.5ATVTPQPRR269 pKa = 11.84TLWTSEE275 pKa = 3.77TGTSGLEE282 pKa = 3.99GPRR285 pKa = 11.84ATRR288 pKa = 11.84VRR290 pKa = 11.84VEE292 pKa = 3.99VLCNIARR299 pKa = 11.84TEE301 pKa = 4.1EE302 pKa = 4.12PQRR305 pKa = 11.84TPTASPDD312 pKa = 3.26ATPVHH317 pKa = 6.74SSTVPEE323 pKa = 4.1EE324 pKa = 4.36TPAGASSVPAPAPVRR339 pKa = 11.84KK340 pKa = 9.46RR341 pKa = 11.84RR342 pKa = 11.84LATEE346 pKa = 4.1NRR348 pKa = 11.84ASSPDD353 pKa = 3.37TRR355 pKa = 11.84GRR357 pKa = 11.84PAADD361 pKa = 3.56PRR363 pKa = 11.84PGTSGAAEE371 pKa = 4.06EE372 pKa = 4.97GEE374 pKa = 4.41EE375 pKa = 4.25EE376 pKa = 4.72DD377 pKa = 5.63GEE379 pKa = 4.93GPTPSDD385 pKa = 3.16PRR387 pKa = 11.84AGRR390 pKa = 11.84DD391 pKa = 3.43QPCRR395 pKa = 11.84KK396 pKa = 9.3CVRR399 pKa = 11.84KK400 pKa = 7.34THH402 pKa = 6.03HH403 pKa = 6.75RR404 pKa = 11.84SALAAEE410 pKa = 4.5RR411 pKa = 11.84PVEE414 pKa = 4.33GSFLQYY420 pKa = 10.45RR421 pKa = 11.84PLVGLSSAVAMAPFLNKK438 pKa = 9.33TVTGDD443 pKa = 3.52CLPIVDD449 pKa = 5.23LEE451 pKa = 4.3SGEE454 pKa = 3.81IGAYY458 pKa = 9.55VVLTHH463 pKa = 6.12QDD465 pKa = 3.2CNMARR470 pKa = 11.84ALAEE474 pKa = 4.36ARR476 pKa = 11.84STWLDD481 pKa = 3.61DD482 pKa = 3.48TSLPLTNPRR491 pKa = 11.84GATGSDD497 pKa = 3.57VPRR500 pKa = 11.84EE501 pKa = 4.11RR502 pKa = 11.84PGSPFRR508 pKa = 11.84LWSTSRR514 pKa = 11.84GTLLFRR520 pKa = 11.84RR521 pKa = 11.84DD522 pKa = 2.94GTLYY526 pKa = 10.65GRR528 pKa = 11.84QSPFGGVQLHH538 pKa = 6.55PWDD541 pKa = 3.83PP542 pKa = 3.55
Molecular weight: 57.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
37452 |
81 |
2844 |
527.5 |
58.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.243 ± 0.255 | 1.784 ± 0.12 |
5.193 ± 0.145 | 4.937 ± 0.156 |
3.917 ± 0.155 | 5.815 ± 0.177 |
2.766 ± 0.092 | 5.281 ± 0.165 |
3.196 ± 0.152 | 10.157 ± 0.24 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.864 ± 0.107 | 3.482 ± 0.124 |
7.847 ± 0.327 | 3.901 ± 0.106 |
6.723 ± 0.199 | 7.148 ± 0.188 |
7.327 ± 0.211 | 6.227 ± 0.138 |
1.143 ± 0.061 | 3.049 ± 0.149 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
