
Human gut microviridae SH-CHD12
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
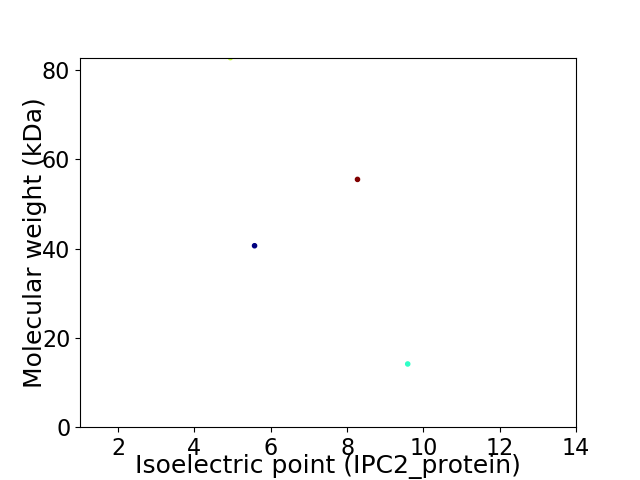
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
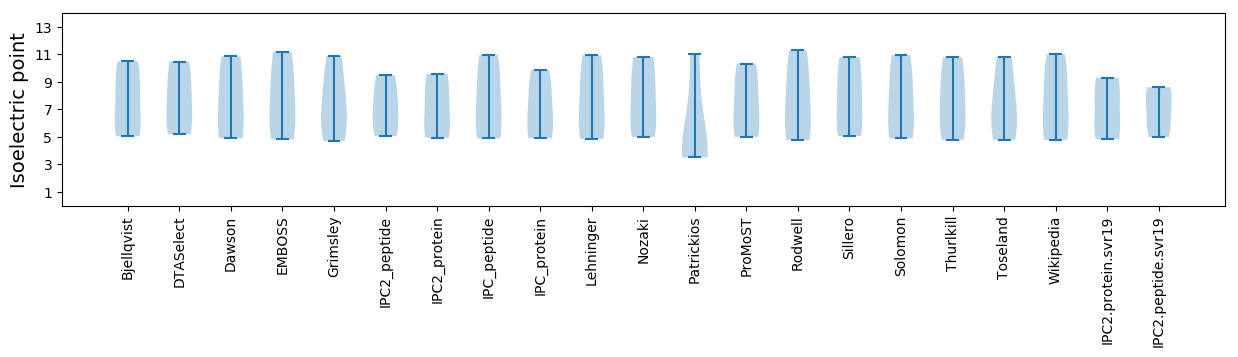
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X9Q116|A0A1X9Q116_9VIRU Putative VP1 OS=Human gut microviridae SH-CHD12 OX=1986032 PE=3 SV=1
MM1 pKa = 7.39IMSNIFSKK9 pKa = 10.57KK10 pKa = 8.87DD11 pKa = 3.27QYY13 pKa = 10.94IDD15 pKa = 3.21RR16 pKa = 11.84VNRR19 pKa = 11.84STFDD23 pKa = 3.37LSFVNNFTTNFGVITPVCLLPVGFGDD49 pKa = 4.7SVQINAKK56 pKa = 10.22FNLQLMPTVFPIQTQMYY73 pKa = 8.47ARR75 pKa = 11.84LHH77 pKa = 5.1FVYY80 pKa = 10.58VRR82 pKa = 11.84TRR84 pKa = 11.84TIWKK88 pKa = 9.14DD89 pKa = 2.92WQKK92 pKa = 10.76FFGGDD97 pKa = 3.32EE98 pKa = 4.33SVVPPFIAFNSQSKK112 pKa = 10.03LDD114 pKa = 4.13AMAQTSTLGDD124 pKa = 3.72YY125 pKa = 10.99LGLPTTIVGKK135 pKa = 8.64YY136 pKa = 9.34GRR138 pKa = 11.84YY139 pKa = 8.98AAYY142 pKa = 8.64PAYY145 pKa = 9.34TPRR148 pKa = 11.84EE149 pKa = 3.83WSYY152 pKa = 11.82VNPLSAIAIALNSVPVDD169 pKa = 3.41ADD171 pKa = 3.58ANALQTALNDD181 pKa = 3.59KK182 pKa = 9.46YY183 pKa = 11.64ASIAAAAPRR192 pKa = 11.84GDD194 pKa = 3.26EE195 pKa = 4.57GIYY198 pKa = 10.18GSAYY202 pKa = 8.81NWYY205 pKa = 10.62AFFTPATGQPFYY217 pKa = 10.65PSQNSDD223 pKa = 2.84NAYY226 pKa = 9.83EE227 pKa = 3.93FSFDD231 pKa = 3.06IDD233 pKa = 3.88FSRR236 pKa = 11.84TSGSLNNTEE245 pKa = 4.37FNNIAFISWAQEE257 pKa = 3.72NGDD260 pKa = 3.75MVVKK264 pKa = 10.29KK265 pKa = 10.47LSNIASASPNGNVTTWNVVIPSSEE289 pKa = 3.72VSGNVIRR296 pKa = 11.84SLALIYY302 pKa = 9.66PKK304 pKa = 10.49NSEE307 pKa = 4.35AIEE310 pKa = 4.07LVNIPANPSYY320 pKa = 10.98NNPWTTNFAFSGFLNVIDD338 pKa = 3.73TTEE341 pKa = 3.85QGKK344 pKa = 8.99VLLDD348 pKa = 3.21KK349 pKa = 11.14CPWYY353 pKa = 10.42DD354 pKa = 3.34GSTEE358 pKa = 4.27SKK360 pKa = 9.0TVRR363 pKa = 11.84ISALPFRR370 pKa = 11.84VYY372 pKa = 9.86EE373 pKa = 4.06AYY375 pKa = 10.99YY376 pKa = 10.28NAFGRR381 pKa = 11.84DD382 pKa = 3.28VRR384 pKa = 11.84NNPFMINGKK393 pKa = 9.8AEE395 pKa = 3.94YY396 pKa = 8.97NQYY399 pKa = 10.51VPTLDD404 pKa = 3.61SGADD408 pKa = 3.39GEE410 pKa = 4.71TYY412 pKa = 10.37EE413 pKa = 4.32LHH415 pKa = 6.1RR416 pKa = 11.84ANWEE420 pKa = 3.52PDD422 pKa = 3.03AYY424 pKa = 8.17TTALQSPQAGVAPLVGITSLGEE446 pKa = 3.7ATFRR450 pKa = 11.84DD451 pKa = 4.3ANGTTYY457 pKa = 9.92TAQLEE462 pKa = 4.35TAEE465 pKa = 5.43DD466 pKa = 4.55GDD468 pKa = 4.59TVTGFQMKK476 pKa = 10.45SSDD479 pKa = 3.42APADD483 pKa = 3.52VVRR486 pKa = 11.84NLIGMATSGISISDD500 pKa = 3.42FRR502 pKa = 11.84NVNSLQRR509 pKa = 11.84FLEE512 pKa = 3.62IRR514 pKa = 11.84IRR516 pKa = 11.84QSPRR520 pKa = 11.84YY521 pKa = 8.62LNLVKK526 pKa = 10.78GLFNVNLDD534 pKa = 3.53YY535 pKa = 11.51DD536 pKa = 4.42EE537 pKa = 6.67LMMPEE542 pKa = 4.27FLGGISDD549 pKa = 3.87NVPVYY554 pKa = 10.47KK555 pKa = 9.71VTQTTPTDD563 pKa = 3.92GSPLGSFAGQGFIQSGMKK581 pKa = 9.93HH582 pKa = 6.43VIRR585 pKa = 11.84KK586 pKa = 6.87YY587 pKa = 10.45CPEE590 pKa = 3.82EE591 pKa = 4.83GYY593 pKa = 10.43ILGVLSVVPAANYY606 pKa = 9.52SQLLAPHH613 pKa = 5.95WTRR616 pKa = 11.84SALLDD621 pKa = 3.1WHH623 pKa = 6.78FPQFNNISFQPMLYY637 pKa = 10.58KK638 pKa = 10.47NLCPLQAYY646 pKa = 8.88NNSPSSLSDD655 pKa = 3.13VYY657 pKa = 10.98GYY659 pKa = 10.31QRR661 pKa = 11.84AWWDD665 pKa = 4.02LISSFDD671 pKa = 3.7EE672 pKa = 3.7VHH674 pKa = 5.63GQFRR678 pKa = 11.84SSLRR682 pKa = 11.84NFVINRR688 pKa = 11.84VFDD691 pKa = 3.86TPPQLSKK698 pKa = 11.38DD699 pKa = 3.77FLLVDD704 pKa = 4.09PAQVNDD710 pKa = 3.62VFAVTAEE717 pKa = 4.15NGDD720 pKa = 4.12KK721 pKa = 10.7ILGSIAFDD729 pKa = 3.16ITKK732 pKa = 8.99KK733 pKa = 6.92TTIPRR738 pKa = 11.84NSIPHH743 pKa = 6.14IEE745 pKa = 3.93
MM1 pKa = 7.39IMSNIFSKK9 pKa = 10.57KK10 pKa = 8.87DD11 pKa = 3.27QYY13 pKa = 10.94IDD15 pKa = 3.21RR16 pKa = 11.84VNRR19 pKa = 11.84STFDD23 pKa = 3.37LSFVNNFTTNFGVITPVCLLPVGFGDD49 pKa = 4.7SVQINAKK56 pKa = 10.22FNLQLMPTVFPIQTQMYY73 pKa = 8.47ARR75 pKa = 11.84LHH77 pKa = 5.1FVYY80 pKa = 10.58VRR82 pKa = 11.84TRR84 pKa = 11.84TIWKK88 pKa = 9.14DD89 pKa = 2.92WQKK92 pKa = 10.76FFGGDD97 pKa = 3.32EE98 pKa = 4.33SVVPPFIAFNSQSKK112 pKa = 10.03LDD114 pKa = 4.13AMAQTSTLGDD124 pKa = 3.72YY125 pKa = 10.99LGLPTTIVGKK135 pKa = 8.64YY136 pKa = 9.34GRR138 pKa = 11.84YY139 pKa = 8.98AAYY142 pKa = 8.64PAYY145 pKa = 9.34TPRR148 pKa = 11.84EE149 pKa = 3.83WSYY152 pKa = 11.82VNPLSAIAIALNSVPVDD169 pKa = 3.41ADD171 pKa = 3.58ANALQTALNDD181 pKa = 3.59KK182 pKa = 9.46YY183 pKa = 11.64ASIAAAAPRR192 pKa = 11.84GDD194 pKa = 3.26EE195 pKa = 4.57GIYY198 pKa = 10.18GSAYY202 pKa = 8.81NWYY205 pKa = 10.62AFFTPATGQPFYY217 pKa = 10.65PSQNSDD223 pKa = 2.84NAYY226 pKa = 9.83EE227 pKa = 3.93FSFDD231 pKa = 3.06IDD233 pKa = 3.88FSRR236 pKa = 11.84TSGSLNNTEE245 pKa = 4.37FNNIAFISWAQEE257 pKa = 3.72NGDD260 pKa = 3.75MVVKK264 pKa = 10.29KK265 pKa = 10.47LSNIASASPNGNVTTWNVVIPSSEE289 pKa = 3.72VSGNVIRR296 pKa = 11.84SLALIYY302 pKa = 9.66PKK304 pKa = 10.49NSEE307 pKa = 4.35AIEE310 pKa = 4.07LVNIPANPSYY320 pKa = 10.98NNPWTTNFAFSGFLNVIDD338 pKa = 3.73TTEE341 pKa = 3.85QGKK344 pKa = 8.99VLLDD348 pKa = 3.21KK349 pKa = 11.14CPWYY353 pKa = 10.42DD354 pKa = 3.34GSTEE358 pKa = 4.27SKK360 pKa = 9.0TVRR363 pKa = 11.84ISALPFRR370 pKa = 11.84VYY372 pKa = 9.86EE373 pKa = 4.06AYY375 pKa = 10.99YY376 pKa = 10.28NAFGRR381 pKa = 11.84DD382 pKa = 3.28VRR384 pKa = 11.84NNPFMINGKK393 pKa = 9.8AEE395 pKa = 3.94YY396 pKa = 8.97NQYY399 pKa = 10.51VPTLDD404 pKa = 3.61SGADD408 pKa = 3.39GEE410 pKa = 4.71TYY412 pKa = 10.37EE413 pKa = 4.32LHH415 pKa = 6.1RR416 pKa = 11.84ANWEE420 pKa = 3.52PDD422 pKa = 3.03AYY424 pKa = 8.17TTALQSPQAGVAPLVGITSLGEE446 pKa = 3.7ATFRR450 pKa = 11.84DD451 pKa = 4.3ANGTTYY457 pKa = 9.92TAQLEE462 pKa = 4.35TAEE465 pKa = 5.43DD466 pKa = 4.55GDD468 pKa = 4.59TVTGFQMKK476 pKa = 10.45SSDD479 pKa = 3.42APADD483 pKa = 3.52VVRR486 pKa = 11.84NLIGMATSGISISDD500 pKa = 3.42FRR502 pKa = 11.84NVNSLQRR509 pKa = 11.84FLEE512 pKa = 3.62IRR514 pKa = 11.84IRR516 pKa = 11.84QSPRR520 pKa = 11.84YY521 pKa = 8.62LNLVKK526 pKa = 10.78GLFNVNLDD534 pKa = 3.53YY535 pKa = 11.51DD536 pKa = 4.42EE537 pKa = 6.67LMMPEE542 pKa = 4.27FLGGISDD549 pKa = 3.87NVPVYY554 pKa = 10.47KK555 pKa = 9.71VTQTTPTDD563 pKa = 3.92GSPLGSFAGQGFIQSGMKK581 pKa = 9.93HH582 pKa = 6.43VIRR585 pKa = 11.84KK586 pKa = 6.87YY587 pKa = 10.45CPEE590 pKa = 3.82EE591 pKa = 4.83GYY593 pKa = 10.43ILGVLSVVPAANYY606 pKa = 9.52SQLLAPHH613 pKa = 5.95WTRR616 pKa = 11.84SALLDD621 pKa = 3.1WHH623 pKa = 6.78FPQFNNISFQPMLYY637 pKa = 10.58KK638 pKa = 10.47NLCPLQAYY646 pKa = 8.88NNSPSSLSDD655 pKa = 3.13VYY657 pKa = 10.98GYY659 pKa = 10.31QRR661 pKa = 11.84AWWDD665 pKa = 4.02LISSFDD671 pKa = 3.7EE672 pKa = 3.7VHH674 pKa = 5.63GQFRR678 pKa = 11.84SSLRR682 pKa = 11.84NFVINRR688 pKa = 11.84VFDD691 pKa = 3.86TPPQLSKK698 pKa = 11.38DD699 pKa = 3.77FLLVDD704 pKa = 4.09PAQVNDD710 pKa = 3.62VFAVTAEE717 pKa = 4.15NGDD720 pKa = 4.12KK721 pKa = 10.7ILGSIAFDD729 pKa = 3.16ITKK732 pKa = 8.99KK733 pKa = 6.92TTIPRR738 pKa = 11.84NSIPHH743 pKa = 6.14IEE745 pKa = 3.93
Molecular weight: 82.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9Q116|A0A1X9Q116_9VIRU Putative VP1 OS=Human gut microviridae SH-CHD12 OX=1986032 PE=3 SV=1
MM1 pKa = 7.58NPVLLLNRR9 pKa = 11.84SFTRR13 pKa = 11.84TNLKK17 pKa = 10.47LRR19 pKa = 11.84MFLPMLSFVSIFLCPPLCRR38 pKa = 11.84LLLLTVRR45 pKa = 11.84FRR47 pKa = 11.84TASLKK52 pKa = 10.42FIKK55 pKa = 10.42LSLKK59 pKa = 7.38MTKK62 pKa = 8.32QHH64 pKa = 6.48LYY66 pKa = 10.47KK67 pKa = 10.62VIEE70 pKa = 4.37VVLTAAISVAAILLLDD86 pKa = 3.89SCSASMSLFWKK97 pKa = 10.14NQGSSQRR104 pKa = 11.84TEE106 pKa = 3.65QSTSARR112 pKa = 11.84VDD114 pKa = 3.49TLKK117 pKa = 11.27SPDD120 pKa = 3.14INLNFF125 pKa = 4.12
MM1 pKa = 7.58NPVLLLNRR9 pKa = 11.84SFTRR13 pKa = 11.84TNLKK17 pKa = 10.47LRR19 pKa = 11.84MFLPMLSFVSIFLCPPLCRR38 pKa = 11.84LLLLTVRR45 pKa = 11.84FRR47 pKa = 11.84TASLKK52 pKa = 10.42FIKK55 pKa = 10.42LSLKK59 pKa = 7.38MTKK62 pKa = 8.32QHH64 pKa = 6.48LYY66 pKa = 10.47KK67 pKa = 10.62VIEE70 pKa = 4.37VVLTAAISVAAILLLDD86 pKa = 3.89SCSASMSLFWKK97 pKa = 10.14NQGSSQRR104 pKa = 11.84TEE106 pKa = 3.65QSTSARR112 pKa = 11.84VDD114 pKa = 3.49TLKK117 pKa = 11.27SPDD120 pKa = 3.14INLNFF125 pKa = 4.12
Molecular weight: 14.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1703 |
125 |
745 |
425.8 |
48.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.34 ± 1.146 | 1.292 ± 0.691 |
5.637 ± 0.349 | 4.11 ± 0.427 |
4.932 ± 0.796 | 5.05 ± 0.754 |
1.527 ± 0.371 | 6.224 ± 0.247 |
4.991 ± 0.844 | 8.28 ± 1.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.29 ± 0.31 | 6.635 ± 0.733 |
4.932 ± 0.699 | 4.815 ± 0.754 |
5.05 ± 0.663 | 8.045 ± 0.916 |
5.755 ± 0.485 | 6.224 ± 0.488 |
1.585 ± 0.115 | 5.285 ± 1.047 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
