
CRESS virus sp. ctf7a5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; unclassified Cressdnaviricota; CRESS viruses
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
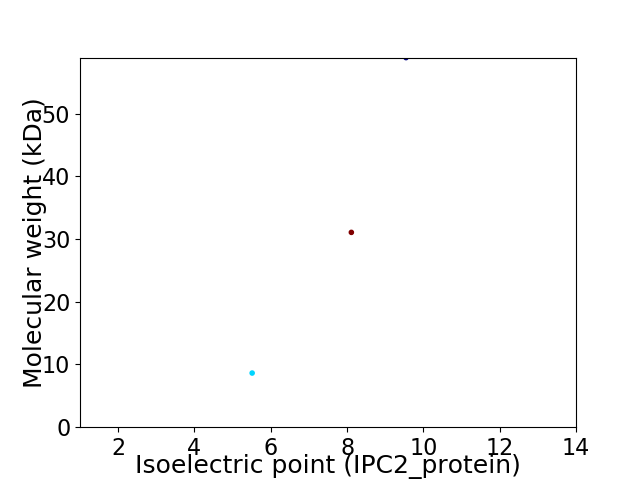
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
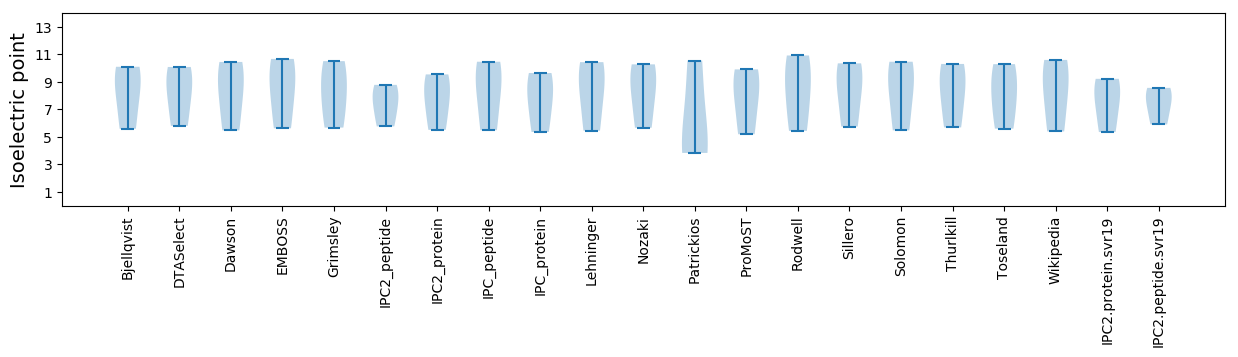
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W902|A0A5Q2W902_9VIRU Putative capsid protein OS=CRESS virus sp. ctf7a5 OX=2656684 PE=4 SV=1
MM1 pKa = 7.93PSTNYY6 pKa = 10.35RR7 pKa = 11.84YY8 pKa = 10.01LSYY11 pKa = 10.69MNYY14 pKa = 10.39LGNKK18 pKa = 7.1WLWIKK23 pKa = 10.89HH24 pKa = 5.82FGDD27 pKa = 3.32TAAQLEE33 pKa = 4.4YY34 pKa = 10.78LDD36 pKa = 5.07DD37 pKa = 5.33HH38 pKa = 9.02IEE40 pKa = 4.03MEE42 pKa = 4.43EE43 pKa = 4.26TFPLTQVRR51 pKa = 11.84SRR53 pKa = 11.84GGNLINEE60 pKa = 4.67LRR62 pKa = 11.84PGSRR66 pKa = 11.84QNPIVIEE73 pKa = 4.07
MM1 pKa = 7.93PSTNYY6 pKa = 10.35RR7 pKa = 11.84YY8 pKa = 10.01LSYY11 pKa = 10.69MNYY14 pKa = 10.39LGNKK18 pKa = 7.1WLWIKK23 pKa = 10.89HH24 pKa = 5.82FGDD27 pKa = 3.32TAAQLEE33 pKa = 4.4YY34 pKa = 10.78LDD36 pKa = 5.07DD37 pKa = 5.33HH38 pKa = 9.02IEE40 pKa = 4.03MEE42 pKa = 4.43EE43 pKa = 4.26TFPLTQVRR51 pKa = 11.84SRR53 pKa = 11.84GGNLINEE60 pKa = 4.67LRR62 pKa = 11.84PGSRR66 pKa = 11.84QNPIVIEE73 pKa = 4.07
Molecular weight: 8.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2WA66|A0A5Q2WA66_9VIRU Replication-associated protein OS=CRESS virus sp. ctf7a5 OX=2656684 PE=4 SV=1
MM1 pKa = 7.73EE2 pKa = 4.46YY3 pKa = 10.68VGGASGGVIGYY14 pKa = 9.95ISGGPWGAYY23 pKa = 9.52SGASLGYY30 pKa = 9.97AAGKK34 pKa = 10.07NFTSNSQMEE43 pKa = 4.39RR44 pKa = 11.84GRR46 pKa = 11.84SRR48 pKa = 11.84VRR50 pKa = 11.84GLTPVRR56 pKa = 11.84VRR58 pKa = 11.84WGSTSRR64 pKa = 11.84EE65 pKa = 4.17RR66 pKa = 11.84KK67 pKa = 8.18WKK69 pKa = 8.82QHH71 pKa = 4.42NLRR74 pKa = 11.84AISRR78 pKa = 11.84QSRR81 pKa = 11.84SWSRR85 pKa = 11.84GRR87 pKa = 11.84EE88 pKa = 3.98LKK90 pKa = 8.83HH91 pKa = 4.45TRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84LGFKK99 pKa = 10.11KK100 pKa = 9.39RR101 pKa = 11.84TVRR104 pKa = 11.84GGGLRR109 pKa = 11.84RR110 pKa = 11.84PVVKK114 pKa = 10.37YY115 pKa = 9.03PKK117 pKa = 9.3RR118 pKa = 11.84SKK120 pKa = 10.9VGFKK124 pKa = 9.93RR125 pKa = 11.84RR126 pKa = 11.84KK127 pKa = 8.89IRR129 pKa = 11.84ISKK132 pKa = 9.12KK133 pKa = 8.47FRR135 pKa = 11.84RR136 pKa = 11.84KK137 pKa = 8.77VMKK140 pKa = 9.99IVTGSKK146 pKa = 10.13YY147 pKa = 10.42KK148 pKa = 10.94GFFMWNTYY156 pKa = 8.61TNVTLATHH164 pKa = 6.48KK165 pKa = 9.74QTVHH169 pKa = 5.48QAQCGFNVQDD179 pKa = 4.49LQNPDD184 pKa = 3.75LGKK187 pKa = 7.48TTYY190 pKa = 10.32PLFSPSVFSAVAEE203 pKa = 4.3TLWGSKK209 pKa = 10.08AGPLSTATSYY219 pKa = 8.86VTGVGNNEE227 pKa = 3.78EE228 pKa = 4.36SNIFSFAKK236 pKa = 10.15IPVINSWVRR245 pKa = 11.84YY246 pKa = 8.63CMKK249 pKa = 10.75NNTSQVLEE257 pKa = 4.06VKK259 pKa = 10.2MFVCKK264 pKa = 10.25PKK266 pKa = 10.17VASFMYY272 pKa = 10.2HH273 pKa = 5.12GMNPAGQSTDD283 pKa = 3.18LAFVYY288 pKa = 10.51DD289 pKa = 6.01PILDD293 pKa = 3.76WEE295 pKa = 4.58KK296 pKa = 11.07AQSDD300 pKa = 4.61LYY302 pKa = 11.19ADD304 pKa = 3.71GVSRR308 pKa = 11.84YY309 pKa = 9.89DD310 pKa = 3.98VLLQKK315 pKa = 9.16QTLKK319 pKa = 10.97GRR321 pKa = 11.84LQTDD325 pKa = 5.31GISTMDD331 pKa = 3.19VDD333 pKa = 4.35PRR335 pKa = 11.84MFTWWNKK342 pKa = 6.99KK343 pKa = 7.34WKK345 pKa = 10.42ADD347 pKa = 3.9VIKK350 pKa = 9.93MKK352 pKa = 10.39IMPGQCIEE360 pKa = 4.95KK361 pKa = 9.79IVKK364 pKa = 10.08GPGDD368 pKa = 3.66FVVDD372 pKa = 3.61TFKK375 pKa = 11.03LWRR378 pKa = 11.84GALEE382 pKa = 4.32NNSSRR387 pKa = 11.84TVEE390 pKa = 4.07NLRR393 pKa = 11.84PAGWTFEE400 pKa = 4.75EE401 pKa = 3.91IQKK404 pKa = 9.28YY405 pKa = 8.56NRR407 pKa = 11.84YY408 pKa = 9.36VFWIVKK414 pKa = 9.25PEE416 pKa = 3.74IASVSTAVVGGVPSAIGVGRR436 pKa = 11.84IPNPEE441 pKa = 3.93GFTGAEE447 pKa = 4.16CFAVEE452 pKa = 3.55QRR454 pKa = 11.84MYY456 pKa = 11.12YY457 pKa = 10.57KK458 pKa = 10.78LALPEE463 pKa = 5.05KK464 pKa = 8.23MGRR467 pKa = 11.84WNEE470 pKa = 3.67VAYY473 pKa = 10.36QSNDD477 pKa = 3.24IISEE481 pKa = 3.97NQMRR485 pKa = 11.84DD486 pKa = 3.24VFFCQYY492 pKa = 10.49SKK494 pKa = 11.01QDD496 pKa = 3.25RR497 pKa = 11.84ANATGSIWQNTQTAGVDD514 pKa = 2.77IFTQIQQ520 pKa = 3.06
MM1 pKa = 7.73EE2 pKa = 4.46YY3 pKa = 10.68VGGASGGVIGYY14 pKa = 9.95ISGGPWGAYY23 pKa = 9.52SGASLGYY30 pKa = 9.97AAGKK34 pKa = 10.07NFTSNSQMEE43 pKa = 4.39RR44 pKa = 11.84GRR46 pKa = 11.84SRR48 pKa = 11.84VRR50 pKa = 11.84GLTPVRR56 pKa = 11.84VRR58 pKa = 11.84WGSTSRR64 pKa = 11.84EE65 pKa = 4.17RR66 pKa = 11.84KK67 pKa = 8.18WKK69 pKa = 8.82QHH71 pKa = 4.42NLRR74 pKa = 11.84AISRR78 pKa = 11.84QSRR81 pKa = 11.84SWSRR85 pKa = 11.84GRR87 pKa = 11.84EE88 pKa = 3.98LKK90 pKa = 8.83HH91 pKa = 4.45TRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84LGFKK99 pKa = 10.11KK100 pKa = 9.39RR101 pKa = 11.84TVRR104 pKa = 11.84GGGLRR109 pKa = 11.84RR110 pKa = 11.84PVVKK114 pKa = 10.37YY115 pKa = 9.03PKK117 pKa = 9.3RR118 pKa = 11.84SKK120 pKa = 10.9VGFKK124 pKa = 9.93RR125 pKa = 11.84RR126 pKa = 11.84KK127 pKa = 8.89IRR129 pKa = 11.84ISKK132 pKa = 9.12KK133 pKa = 8.47FRR135 pKa = 11.84RR136 pKa = 11.84KK137 pKa = 8.77VMKK140 pKa = 9.99IVTGSKK146 pKa = 10.13YY147 pKa = 10.42KK148 pKa = 10.94GFFMWNTYY156 pKa = 8.61TNVTLATHH164 pKa = 6.48KK165 pKa = 9.74QTVHH169 pKa = 5.48QAQCGFNVQDD179 pKa = 4.49LQNPDD184 pKa = 3.75LGKK187 pKa = 7.48TTYY190 pKa = 10.32PLFSPSVFSAVAEE203 pKa = 4.3TLWGSKK209 pKa = 10.08AGPLSTATSYY219 pKa = 8.86VTGVGNNEE227 pKa = 3.78EE228 pKa = 4.36SNIFSFAKK236 pKa = 10.15IPVINSWVRR245 pKa = 11.84YY246 pKa = 8.63CMKK249 pKa = 10.75NNTSQVLEE257 pKa = 4.06VKK259 pKa = 10.2MFVCKK264 pKa = 10.25PKK266 pKa = 10.17VASFMYY272 pKa = 10.2HH273 pKa = 5.12GMNPAGQSTDD283 pKa = 3.18LAFVYY288 pKa = 10.51DD289 pKa = 6.01PILDD293 pKa = 3.76WEE295 pKa = 4.58KK296 pKa = 11.07AQSDD300 pKa = 4.61LYY302 pKa = 11.19ADD304 pKa = 3.71GVSRR308 pKa = 11.84YY309 pKa = 9.89DD310 pKa = 3.98VLLQKK315 pKa = 9.16QTLKK319 pKa = 10.97GRR321 pKa = 11.84LQTDD325 pKa = 5.31GISTMDD331 pKa = 3.19VDD333 pKa = 4.35PRR335 pKa = 11.84MFTWWNKK342 pKa = 6.99KK343 pKa = 7.34WKK345 pKa = 10.42ADD347 pKa = 3.9VIKK350 pKa = 9.93MKK352 pKa = 10.39IMPGQCIEE360 pKa = 4.95KK361 pKa = 9.79IVKK364 pKa = 10.08GPGDD368 pKa = 3.66FVVDD372 pKa = 3.61TFKK375 pKa = 11.03LWRR378 pKa = 11.84GALEE382 pKa = 4.32NNSSRR387 pKa = 11.84TVEE390 pKa = 4.07NLRR393 pKa = 11.84PAGWTFEE400 pKa = 4.75EE401 pKa = 3.91IQKK404 pKa = 9.28YY405 pKa = 8.56NRR407 pKa = 11.84YY408 pKa = 9.36VFWIVKK414 pKa = 9.25PEE416 pKa = 3.74IASVSTAVVGGVPSAIGVGRR436 pKa = 11.84IPNPEE441 pKa = 3.93GFTGAEE447 pKa = 4.16CFAVEE452 pKa = 3.55QRR454 pKa = 11.84MYY456 pKa = 11.12YY457 pKa = 10.57KK458 pKa = 10.78LALPEE463 pKa = 5.05KK464 pKa = 8.23MGRR467 pKa = 11.84WNEE470 pKa = 3.67VAYY473 pKa = 10.36QSNDD477 pKa = 3.24IISEE481 pKa = 3.97NQMRR485 pKa = 11.84DD486 pKa = 3.24VFFCQYY492 pKa = 10.49SKK494 pKa = 11.01QDD496 pKa = 3.25RR497 pKa = 11.84ANATGSIWQNTQTAGVDD514 pKa = 2.77IFTQIQQ520 pKa = 3.06
Molecular weight: 58.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
861 |
73 |
520 |
287.0 |
32.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.039 ± 0.798 | 1.045 ± 0.235 |
4.065 ± 0.527 | 5.343 ± 1.14 |
3.717 ± 0.752 | 8.246 ± 0.674 |
1.51 ± 0.481 | 4.878 ± 0.448 |
6.62 ± 1.205 | 6.852 ± 1.763 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.671 ± 0.433 | 4.53 ± 0.968 |
4.53 ± 0.421 | 3.949 ± 0.766 |
7.782 ± 0.376 | 6.736 ± 0.801 |
5.923 ± 0.198 | 7.549 ± 1.171 |
3.484 ± 0.443 | 4.53 ± 0.678 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
