
Mycoplasma virus P1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Delislevirus
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
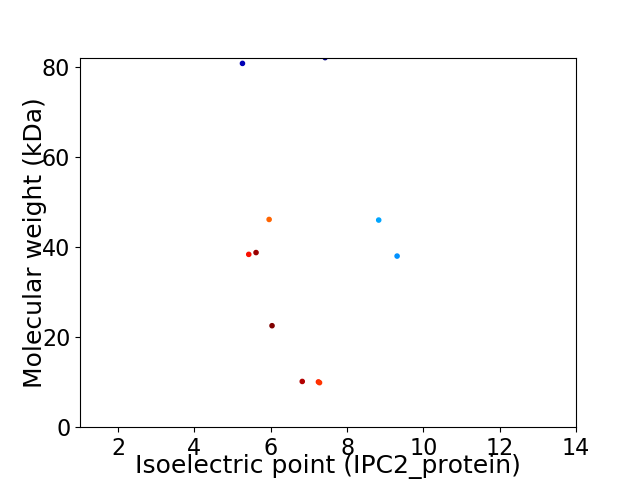
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q9FZR3|Q9FZR3_9CAUD p23 OS=Mycoplasma virus P1 OX=35238 GN=orf6 PE=4 SV=1
MM1 pKa = 7.8LYY3 pKa = 10.75YY4 pKa = 10.2IFNSDD9 pKa = 2.88VNYY12 pKa = 10.46FDD14 pKa = 3.96NPKK17 pKa = 10.75NKK19 pKa = 8.53TLRR22 pKa = 11.84QWILRR27 pKa = 11.84HH28 pKa = 5.89FNTSLQGSFDD38 pKa = 4.13LSWAQEE44 pKa = 4.04RR45 pKa = 11.84FFSFQSDD52 pKa = 3.83QEE54 pKa = 4.4VKK56 pKa = 10.5RR57 pKa = 11.84LFLISPTNDD66 pKa = 2.99KK67 pKa = 10.91NDD69 pKa = 3.76PKK71 pKa = 11.33AQICYY76 pKa = 9.93VNNIEE81 pKa = 5.42YY82 pKa = 10.65LDD84 pKa = 3.54SDD86 pKa = 4.11KK87 pKa = 11.28NLKK90 pKa = 10.12RR91 pKa = 11.84IYY93 pKa = 10.96AEE95 pKa = 3.46VDD97 pKa = 2.98SFYY100 pKa = 10.81DD101 pKa = 4.25INIDD105 pKa = 4.94DD106 pKa = 4.39IGGKK110 pKa = 9.46LKK112 pKa = 9.81RR113 pKa = 11.84TNNWLLFEE121 pKa = 4.62GKK123 pKa = 7.66YY124 pKa = 10.38HH125 pKa = 7.02KK126 pKa = 10.63NLLALEE132 pKa = 5.11GINLPKK138 pKa = 10.28LDD140 pKa = 3.81FEE142 pKa = 4.88KK143 pKa = 11.05VVTLDD148 pKa = 3.18YY149 pKa = 11.11YY150 pKa = 9.87KK151 pKa = 10.79QKK153 pKa = 10.79DD154 pKa = 3.19ISFQIRR160 pKa = 11.84DD161 pKa = 3.23KK162 pKa = 11.07AIRR165 pKa = 11.84LEE167 pKa = 3.87RR168 pKa = 11.84DD169 pKa = 3.31YY170 pKa = 11.4LIKK173 pKa = 10.89DD174 pKa = 3.26NNFIRR179 pKa = 11.84KK180 pKa = 8.16PLQYY184 pKa = 11.02SLDD187 pKa = 3.58FLEE190 pKa = 5.97RR191 pKa = 11.84IEE193 pKa = 5.03DD194 pKa = 3.7LQTRR198 pKa = 11.84EE199 pKa = 3.77MLEE202 pKa = 3.91AQEE205 pKa = 4.14EE206 pKa = 4.6NFWFWQNNQRR216 pKa = 11.84VPDD219 pKa = 4.09SIINHH224 pKa = 6.15FKK226 pKa = 10.87NNQSFQRR233 pKa = 11.84YY234 pKa = 7.9SVSDD238 pKa = 3.35QEE240 pKa = 4.67IINFINRR247 pKa = 11.84WYY249 pKa = 10.3DD250 pKa = 3.13RR251 pKa = 11.84WIFWLPTSTSTFIVISEE268 pKa = 4.29KK269 pKa = 10.71KK270 pKa = 9.25VVAINFLFTKK280 pKa = 9.7RR281 pKa = 11.84TYY283 pKa = 11.22NEE285 pKa = 4.1GNPPNHH291 pKa = 6.63RR292 pKa = 11.84LVEE295 pKa = 4.31RR296 pKa = 11.84NTIDD300 pKa = 3.65YY301 pKa = 10.17SEE303 pKa = 4.17QSIASEE309 pKa = 4.16EE310 pKa = 4.07WSDD313 pKa = 4.26LIFWKK318 pKa = 10.71SLAIVGLPEE327 pKa = 3.64IKK329 pKa = 10.41GAFVSWLKK337 pKa = 10.86EE338 pKa = 3.83KK339 pKa = 10.83AHH341 pKa = 6.67LNLEE345 pKa = 4.43PTFNTYY351 pKa = 11.2NDD353 pKa = 2.91VKK355 pKa = 10.41TFIEE359 pKa = 4.48EE360 pKa = 4.23KK361 pKa = 10.46FKK363 pKa = 11.66SNANGFLYY371 pKa = 10.44KK372 pKa = 9.3PTQEE376 pKa = 5.54LIIDD380 pKa = 4.31FNNLEE385 pKa = 4.03PLMNKK390 pKa = 8.31EE391 pKa = 3.57WTTLTFSKK399 pKa = 9.95KK400 pKa = 10.03HH401 pKa = 5.95KK402 pKa = 9.67IALSKK407 pKa = 10.9EE408 pKa = 3.75FVEE411 pKa = 4.48YY412 pKa = 10.58QVISPNCTMIIDD424 pKa = 4.02YY425 pKa = 9.07EE426 pKa = 4.27QVVAKK431 pKa = 10.61EE432 pKa = 4.17NFNDD436 pKa = 4.13SIRR439 pKa = 11.84FRR441 pKa = 11.84YY442 pKa = 9.74SEE444 pKa = 4.56IEE446 pKa = 4.05TQEE449 pKa = 3.55AGYY452 pKa = 10.34CILEE456 pKa = 4.18VEE458 pKa = 4.52GQDD461 pKa = 3.5FRR463 pKa = 11.84QVQSYY468 pKa = 10.02LKK470 pKa = 10.56NDD472 pKa = 3.51VLITNAYY479 pKa = 9.29KK480 pKa = 10.62SHH482 pKa = 7.43LDD484 pKa = 3.77ANWLSHH490 pKa = 5.41QFSSQSLQIAKK501 pKa = 9.51QRR503 pKa = 11.84ADD505 pKa = 3.55NDD507 pKa = 3.62FLLNTIKK514 pKa = 10.81NAIALPFSAISSFGSTFQGGSVVAGLNAIGGVINTGFNFASSIIGGLRR562 pKa = 11.84AQEE565 pKa = 3.96DD566 pKa = 3.79AQRR569 pKa = 11.84AIEE572 pKa = 4.64GFNATYY578 pKa = 10.83KK579 pKa = 10.57DD580 pKa = 3.74LQSQEE585 pKa = 4.54RR586 pKa = 11.84FLSNNTSLTSLLINKK601 pKa = 8.81EE602 pKa = 3.96NEE604 pKa = 3.41IALYY608 pKa = 9.4IAKK611 pKa = 9.55PVEE614 pKa = 3.97KK615 pKa = 10.33DD616 pKa = 3.14LYY618 pKa = 10.67ALNRR622 pKa = 11.84YY623 pKa = 7.91FQDD626 pKa = 3.68FGQSVNDD633 pKa = 3.67YY634 pKa = 10.72YY635 pKa = 11.51SYY637 pKa = 11.62SLYY640 pKa = 10.87NINEE644 pKa = 3.93ALKK647 pKa = 9.6FSIYY651 pKa = 10.9GLYY654 pKa = 10.31NQFEE658 pKa = 4.82YY659 pKa = 10.61IEE661 pKa = 5.52DD662 pKa = 3.63IPNVSWEE669 pKa = 4.01VNNYY673 pKa = 9.76IKK675 pKa = 10.85DD676 pKa = 3.61LAKK679 pKa = 9.8TGFWIKK685 pKa = 10.63KK686 pKa = 8.76YY687 pKa = 10.54DD688 pKa = 3.33
MM1 pKa = 7.8LYY3 pKa = 10.75YY4 pKa = 10.2IFNSDD9 pKa = 2.88VNYY12 pKa = 10.46FDD14 pKa = 3.96NPKK17 pKa = 10.75NKK19 pKa = 8.53TLRR22 pKa = 11.84QWILRR27 pKa = 11.84HH28 pKa = 5.89FNTSLQGSFDD38 pKa = 4.13LSWAQEE44 pKa = 4.04RR45 pKa = 11.84FFSFQSDD52 pKa = 3.83QEE54 pKa = 4.4VKK56 pKa = 10.5RR57 pKa = 11.84LFLISPTNDD66 pKa = 2.99KK67 pKa = 10.91NDD69 pKa = 3.76PKK71 pKa = 11.33AQICYY76 pKa = 9.93VNNIEE81 pKa = 5.42YY82 pKa = 10.65LDD84 pKa = 3.54SDD86 pKa = 4.11KK87 pKa = 11.28NLKK90 pKa = 10.12RR91 pKa = 11.84IYY93 pKa = 10.96AEE95 pKa = 3.46VDD97 pKa = 2.98SFYY100 pKa = 10.81DD101 pKa = 4.25INIDD105 pKa = 4.94DD106 pKa = 4.39IGGKK110 pKa = 9.46LKK112 pKa = 9.81RR113 pKa = 11.84TNNWLLFEE121 pKa = 4.62GKK123 pKa = 7.66YY124 pKa = 10.38HH125 pKa = 7.02KK126 pKa = 10.63NLLALEE132 pKa = 5.11GINLPKK138 pKa = 10.28LDD140 pKa = 3.81FEE142 pKa = 4.88KK143 pKa = 11.05VVTLDD148 pKa = 3.18YY149 pKa = 11.11YY150 pKa = 9.87KK151 pKa = 10.79QKK153 pKa = 10.79DD154 pKa = 3.19ISFQIRR160 pKa = 11.84DD161 pKa = 3.23KK162 pKa = 11.07AIRR165 pKa = 11.84LEE167 pKa = 3.87RR168 pKa = 11.84DD169 pKa = 3.31YY170 pKa = 11.4LIKK173 pKa = 10.89DD174 pKa = 3.26NNFIRR179 pKa = 11.84KK180 pKa = 8.16PLQYY184 pKa = 11.02SLDD187 pKa = 3.58FLEE190 pKa = 5.97RR191 pKa = 11.84IEE193 pKa = 5.03DD194 pKa = 3.7LQTRR198 pKa = 11.84EE199 pKa = 3.77MLEE202 pKa = 3.91AQEE205 pKa = 4.14EE206 pKa = 4.6NFWFWQNNQRR216 pKa = 11.84VPDD219 pKa = 4.09SIINHH224 pKa = 6.15FKK226 pKa = 10.87NNQSFQRR233 pKa = 11.84YY234 pKa = 7.9SVSDD238 pKa = 3.35QEE240 pKa = 4.67IINFINRR247 pKa = 11.84WYY249 pKa = 10.3DD250 pKa = 3.13RR251 pKa = 11.84WIFWLPTSTSTFIVISEE268 pKa = 4.29KK269 pKa = 10.71KK270 pKa = 9.25VVAINFLFTKK280 pKa = 9.7RR281 pKa = 11.84TYY283 pKa = 11.22NEE285 pKa = 4.1GNPPNHH291 pKa = 6.63RR292 pKa = 11.84LVEE295 pKa = 4.31RR296 pKa = 11.84NTIDD300 pKa = 3.65YY301 pKa = 10.17SEE303 pKa = 4.17QSIASEE309 pKa = 4.16EE310 pKa = 4.07WSDD313 pKa = 4.26LIFWKK318 pKa = 10.71SLAIVGLPEE327 pKa = 3.64IKK329 pKa = 10.41GAFVSWLKK337 pKa = 10.86EE338 pKa = 3.83KK339 pKa = 10.83AHH341 pKa = 6.67LNLEE345 pKa = 4.43PTFNTYY351 pKa = 11.2NDD353 pKa = 2.91VKK355 pKa = 10.41TFIEE359 pKa = 4.48EE360 pKa = 4.23KK361 pKa = 10.46FKK363 pKa = 11.66SNANGFLYY371 pKa = 10.44KK372 pKa = 9.3PTQEE376 pKa = 5.54LIIDD380 pKa = 4.31FNNLEE385 pKa = 4.03PLMNKK390 pKa = 8.31EE391 pKa = 3.57WTTLTFSKK399 pKa = 9.95KK400 pKa = 10.03HH401 pKa = 5.95KK402 pKa = 9.67IALSKK407 pKa = 10.9EE408 pKa = 3.75FVEE411 pKa = 4.48YY412 pKa = 10.58QVISPNCTMIIDD424 pKa = 4.02YY425 pKa = 9.07EE426 pKa = 4.27QVVAKK431 pKa = 10.61EE432 pKa = 4.17NFNDD436 pKa = 4.13SIRR439 pKa = 11.84FRR441 pKa = 11.84YY442 pKa = 9.74SEE444 pKa = 4.56IEE446 pKa = 4.05TQEE449 pKa = 3.55AGYY452 pKa = 10.34CILEE456 pKa = 4.18VEE458 pKa = 4.52GQDD461 pKa = 3.5FRR463 pKa = 11.84QVQSYY468 pKa = 10.02LKK470 pKa = 10.56NDD472 pKa = 3.51VLITNAYY479 pKa = 9.29KK480 pKa = 10.62SHH482 pKa = 7.43LDD484 pKa = 3.77ANWLSHH490 pKa = 5.41QFSSQSLQIAKK501 pKa = 9.51QRR503 pKa = 11.84ADD505 pKa = 3.55NDD507 pKa = 3.62FLLNTIKK514 pKa = 10.81NAIALPFSAISSFGSTFQGGSVVAGLNAIGGVINTGFNFASSIIGGLRR562 pKa = 11.84AQEE565 pKa = 3.96DD566 pKa = 3.79AQRR569 pKa = 11.84AIEE572 pKa = 4.64GFNATYY578 pKa = 10.83KK579 pKa = 10.57DD580 pKa = 3.74LQSQEE585 pKa = 4.54RR586 pKa = 11.84FLSNNTSLTSLLINKK601 pKa = 8.81EE602 pKa = 3.96NEE604 pKa = 3.41IALYY608 pKa = 9.4IAKK611 pKa = 9.55PVEE614 pKa = 3.97KK615 pKa = 10.33DD616 pKa = 3.14LYY618 pKa = 10.67ALNRR622 pKa = 11.84YY623 pKa = 7.91FQDD626 pKa = 3.68FGQSVNDD633 pKa = 3.67YY634 pKa = 10.72YY635 pKa = 11.51SYY637 pKa = 11.62SLYY640 pKa = 10.87NINEE644 pKa = 3.93ALKK647 pKa = 9.6FSIYY651 pKa = 10.9GLYY654 pKa = 10.31NQFEE658 pKa = 4.82YY659 pKa = 10.61IEE661 pKa = 5.52DD662 pKa = 3.63IPNVSWEE669 pKa = 4.01VNNYY673 pKa = 9.76IKK675 pKa = 10.85DD676 pKa = 3.61LAKK679 pKa = 9.8TGFWIKK685 pKa = 10.63KK686 pKa = 8.76YY687 pKa = 10.54DD688 pKa = 3.33
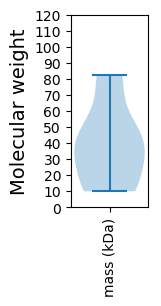
Molecular weight: 80.9 kDa
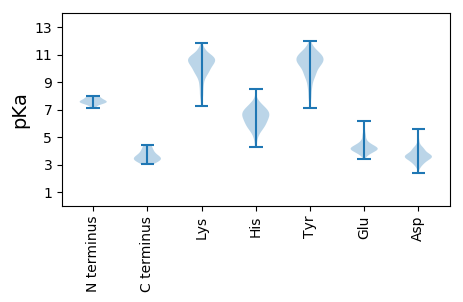
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9FZR8|Q9FZR8_9CAUD DNA-directed DNA polymerase OS=Mycoplasma virus P1 OX=35238 GN=orf1 PE=3 SV=1
MM1 pKa = 7.69SRR3 pKa = 11.84RR4 pKa = 11.84DD5 pKa = 3.75KK6 pKa = 11.56LKK8 pKa = 9.91DD9 pKa = 3.06TYY11 pKa = 11.73KK12 pKa = 10.79LFKK15 pKa = 10.66DD16 pKa = 3.5DD17 pKa = 5.6FKK19 pKa = 11.83ALGYY23 pKa = 9.54SQNKK27 pKa = 7.51FVKK30 pKa = 9.08EE31 pKa = 4.1FAHH34 pKa = 5.73VRR36 pKa = 11.84YY37 pKa = 9.54YY38 pKa = 11.02DD39 pKa = 3.54KK40 pKa = 10.97KK41 pKa = 10.4KK42 pKa = 10.89KK43 pKa = 8.3EE44 pKa = 4.0LKK46 pKa = 9.68ISKK49 pKa = 9.89INNRR53 pKa = 11.84LIAKK57 pKa = 9.73RR58 pKa = 11.84EE59 pKa = 3.67RR60 pKa = 11.84YY61 pKa = 8.0YY62 pKa = 11.19QKK64 pKa = 10.23LTTTSKK70 pKa = 10.81KK71 pKa = 10.35QINEE75 pKa = 3.83NLTKK79 pKa = 10.22EE80 pKa = 4.49FSNIAHH86 pKa = 6.99FSLKK90 pKa = 10.33ARR92 pKa = 11.84NLKK95 pKa = 7.99EE96 pKa = 4.16TQLKK100 pKa = 10.92SNIKK104 pKa = 8.5NTLNTLEE111 pKa = 4.21KK112 pKa = 10.51YY113 pKa = 10.15IRR115 pKa = 11.84TEE117 pKa = 3.77EE118 pKa = 3.82DD119 pKa = 2.53RR120 pKa = 11.84KK121 pKa = 10.71RR122 pKa = 11.84LFNIKK127 pKa = 10.03FVVNHH132 pKa = 6.99KK133 pKa = 9.21DD134 pKa = 3.39QKK136 pKa = 10.67VMNGLAFVNYY146 pKa = 7.84LTNHH150 pKa = 6.36GKK152 pKa = 10.18LDD154 pKa = 4.0SYY156 pKa = 10.57PFNYY160 pKa = 10.19SNEE163 pKa = 4.23QIKK166 pKa = 10.62SATKK170 pKa = 9.74ALRR173 pKa = 11.84YY174 pKa = 9.08VLKK177 pKa = 11.01NKK179 pKa = 9.74IDD181 pKa = 3.9FNSTKK186 pKa = 10.94DD187 pKa = 3.21FTKK190 pKa = 10.92YY191 pKa = 10.77FNTSLNKK198 pKa = 10.0QEE200 pKa = 4.15IKK202 pKa = 10.57KK203 pKa = 10.06HH204 pKa = 4.24GFKK207 pKa = 10.22TFSASEE213 pKa = 4.45VYY215 pKa = 10.38GVAGSTFNNLGFLRR229 pKa = 11.84VEE231 pKa = 3.98LTTYY235 pKa = 10.69DD236 pKa = 4.5SKK238 pKa = 11.34FQSQRR243 pKa = 11.84VNNVEE248 pKa = 3.98LVGEE252 pKa = 4.22SSKK255 pKa = 11.58ALIKK259 pKa = 9.27WAAYY263 pKa = 9.53LKK265 pKa = 10.57QKK267 pKa = 10.57GINQFLIDD275 pKa = 4.22AFGVDD280 pKa = 3.31IAEE283 pKa = 4.14NMHH286 pKa = 6.99LYY288 pKa = 10.27GPKK291 pKa = 8.71NSKK294 pKa = 8.86WLFVNEE300 pKa = 4.12KK301 pKa = 8.76DD302 pKa = 3.18HH303 pKa = 7.27EE304 pKa = 4.53YY305 pKa = 11.02FFFYY309 pKa = 10.55FNSAKK314 pKa = 10.15EE315 pKa = 3.63IYY317 pKa = 9.14YY318 pKa = 10.83ANRR321 pKa = 3.4
MM1 pKa = 7.69SRR3 pKa = 11.84RR4 pKa = 11.84DD5 pKa = 3.75KK6 pKa = 11.56LKK8 pKa = 9.91DD9 pKa = 3.06TYY11 pKa = 11.73KK12 pKa = 10.79LFKK15 pKa = 10.66DD16 pKa = 3.5DD17 pKa = 5.6FKK19 pKa = 11.83ALGYY23 pKa = 9.54SQNKK27 pKa = 7.51FVKK30 pKa = 9.08EE31 pKa = 4.1FAHH34 pKa = 5.73VRR36 pKa = 11.84YY37 pKa = 9.54YY38 pKa = 11.02DD39 pKa = 3.54KK40 pKa = 10.97KK41 pKa = 10.4KK42 pKa = 10.89KK43 pKa = 8.3EE44 pKa = 4.0LKK46 pKa = 9.68ISKK49 pKa = 9.89INNRR53 pKa = 11.84LIAKK57 pKa = 9.73RR58 pKa = 11.84EE59 pKa = 3.67RR60 pKa = 11.84YY61 pKa = 8.0YY62 pKa = 11.19QKK64 pKa = 10.23LTTTSKK70 pKa = 10.81KK71 pKa = 10.35QINEE75 pKa = 3.83NLTKK79 pKa = 10.22EE80 pKa = 4.49FSNIAHH86 pKa = 6.99FSLKK90 pKa = 10.33ARR92 pKa = 11.84NLKK95 pKa = 7.99EE96 pKa = 4.16TQLKK100 pKa = 10.92SNIKK104 pKa = 8.5NTLNTLEE111 pKa = 4.21KK112 pKa = 10.51YY113 pKa = 10.15IRR115 pKa = 11.84TEE117 pKa = 3.77EE118 pKa = 3.82DD119 pKa = 2.53RR120 pKa = 11.84KK121 pKa = 10.71RR122 pKa = 11.84LFNIKK127 pKa = 10.03FVVNHH132 pKa = 6.99KK133 pKa = 9.21DD134 pKa = 3.39QKK136 pKa = 10.67VMNGLAFVNYY146 pKa = 7.84LTNHH150 pKa = 6.36GKK152 pKa = 10.18LDD154 pKa = 4.0SYY156 pKa = 10.57PFNYY160 pKa = 10.19SNEE163 pKa = 4.23QIKK166 pKa = 10.62SATKK170 pKa = 9.74ALRR173 pKa = 11.84YY174 pKa = 9.08VLKK177 pKa = 11.01NKK179 pKa = 9.74IDD181 pKa = 3.9FNSTKK186 pKa = 10.94DD187 pKa = 3.21FTKK190 pKa = 10.92YY191 pKa = 10.77FNTSLNKK198 pKa = 10.0QEE200 pKa = 4.15IKK202 pKa = 10.57KK203 pKa = 10.06HH204 pKa = 4.24GFKK207 pKa = 10.22TFSASEE213 pKa = 4.45VYY215 pKa = 10.38GVAGSTFNNLGFLRR229 pKa = 11.84VEE231 pKa = 3.98LTTYY235 pKa = 10.69DD236 pKa = 4.5SKK238 pKa = 11.34FQSQRR243 pKa = 11.84VNNVEE248 pKa = 3.98LVGEE252 pKa = 4.22SSKK255 pKa = 11.58ALIKK259 pKa = 9.27WAAYY263 pKa = 9.53LKK265 pKa = 10.57QKK267 pKa = 10.57GINQFLIDD275 pKa = 4.22AFGVDD280 pKa = 3.31IAEE283 pKa = 4.14NMHH286 pKa = 6.99LYY288 pKa = 10.27GPKK291 pKa = 8.71NSKK294 pKa = 8.86WLFVNEE300 pKa = 4.12KK301 pKa = 8.76DD302 pKa = 3.18HH303 pKa = 7.27EE304 pKa = 4.53YY305 pKa = 11.02FFFYY309 pKa = 10.55FNSAKK314 pKa = 10.15EE315 pKa = 3.63IYY317 pKa = 9.14YY318 pKa = 10.83ANRR321 pKa = 3.4
Molecular weight: 38.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3579 |
80 |
694 |
325.4 |
38.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.744 ± 0.544 | 0.363 ± 0.154 |
5.979 ± 0.376 | 7.376 ± 0.509 |
6.706 ± 0.277 | 3.325 ± 0.391 |
1.565 ± 0.224 | 8.243 ± 0.502 |
10.198 ± 0.965 | 9.36 ± 0.325 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.76 ± 0.335 | 9.193 ± 0.898 |
2.012 ± 0.269 | 4.219 ± 0.349 |
3.744 ± 0.26 | 6.454 ± 0.474 |
4.722 ± 0.302 | 4.331 ± 0.234 |
1.146 ± 0.306 | 5.56 ± 0.458 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
