
Avon-Heathcote Estuary associated circular virus 7
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.72
Get precalculated fractions of proteins
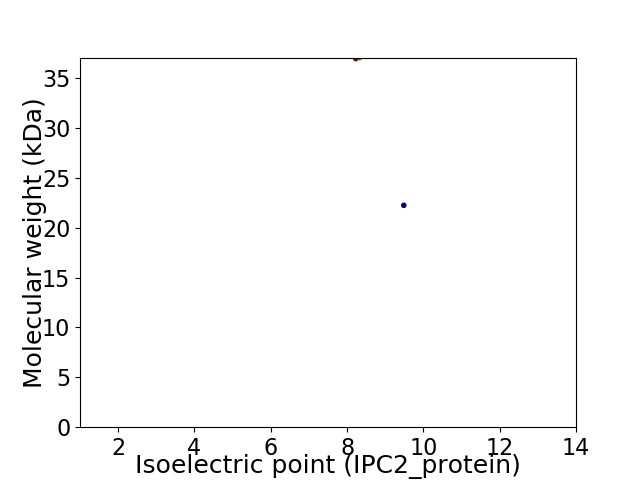
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IAX7|A0A0C5IAX7_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 7 OX=1618258 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.3KK3 pKa = 10.14FKK5 pKa = 10.94EE6 pKa = 4.47KK7 pKa = 9.33ITSLLMRR14 pKa = 11.84FRR16 pKa = 11.84NVVGTLFTNEE26 pKa = 3.99RR27 pKa = 11.84PHH29 pKa = 5.96EE30 pKa = 4.01QFITSPLIKK39 pKa = 10.48YY40 pKa = 9.06FIGQYY45 pKa = 8.71EE46 pKa = 4.12ICPSSKK52 pKa = 9.6RR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.99LQFYY59 pKa = 10.39CEE61 pKa = 4.38FVGQQRR67 pKa = 11.84LGAVRR72 pKa = 11.84QLFGPTVHH80 pKa = 6.09VEE82 pKa = 4.03PRR84 pKa = 11.84RR85 pKa = 11.84GTVDD89 pKa = 3.19EE90 pKa = 4.36ARR92 pKa = 11.84TYY94 pKa = 10.65CSKK97 pKa = 10.99EE98 pKa = 3.87DD99 pKa = 3.57TRR101 pKa = 11.84EE102 pKa = 3.88PGIDD106 pKa = 3.14SGPFEE111 pKa = 5.14NGTPSKK117 pKa = 10.4SGKK120 pKa = 10.11RR121 pKa = 11.84SDD123 pKa = 3.55LADD126 pKa = 3.28IKK128 pKa = 11.22EE129 pKa = 4.06EE130 pKa = 4.05LDD132 pKa = 3.52AGSSLKK138 pKa = 10.81SISQQHH144 pKa = 6.14FGQFLRR150 pKa = 11.84YY151 pKa = 8.7RR152 pKa = 11.84KK153 pKa = 10.06SFEE156 pKa = 3.78AYY158 pKa = 8.89IVLNQDD164 pKa = 2.48PRR166 pKa = 11.84TWEE169 pKa = 3.96MEE171 pKa = 3.69NSILWGEE178 pKa = 4.2PGTGKK183 pKa = 8.67TKK185 pKa = 10.48LAYY188 pKa = 9.63DD189 pKa = 3.9LKK191 pKa = 10.99EE192 pKa = 4.03RR193 pKa = 11.84DD194 pKa = 3.72GVEE197 pKa = 4.38GYY199 pKa = 10.02PLMRR203 pKa = 11.84NQNGNVWFDD212 pKa = 3.68GYY214 pKa = 10.71HH215 pKa = 4.96GQEE218 pKa = 3.76ILLIDD223 pKa = 5.28DD224 pKa = 3.98YY225 pKa = 11.69YY226 pKa = 11.84GWIPLAFLLQLLDD239 pKa = 4.24RR240 pKa = 11.84YY241 pKa = 9.89PMNVQTKK248 pKa = 8.54GGSVPFTSKK257 pKa = 10.81KK258 pKa = 10.41IIITSNKK265 pKa = 9.87SPEE268 pKa = 3.74CWYY271 pKa = 10.48NWSKK275 pKa = 10.69FGKK278 pKa = 9.96NMFGAFEE285 pKa = 3.88RR286 pKa = 11.84RR287 pKa = 11.84INQVFHH293 pKa = 5.35YY294 pKa = 10.83VKK296 pKa = 10.78DD297 pKa = 3.82KK298 pKa = 11.43DD299 pKa = 3.52PAIYY303 pKa = 9.13PIKK306 pKa = 10.54FPEE309 pKa = 4.26TVSAVEE315 pKa = 4.46FFQQ318 pKa = 5.57
MM1 pKa = 7.61KK2 pKa = 10.3KK3 pKa = 10.14FKK5 pKa = 10.94EE6 pKa = 4.47KK7 pKa = 9.33ITSLLMRR14 pKa = 11.84FRR16 pKa = 11.84NVVGTLFTNEE26 pKa = 3.99RR27 pKa = 11.84PHH29 pKa = 5.96EE30 pKa = 4.01QFITSPLIKK39 pKa = 10.48YY40 pKa = 9.06FIGQYY45 pKa = 8.71EE46 pKa = 4.12ICPSSKK52 pKa = 9.6RR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.99LQFYY59 pKa = 10.39CEE61 pKa = 4.38FVGQQRR67 pKa = 11.84LGAVRR72 pKa = 11.84QLFGPTVHH80 pKa = 6.09VEE82 pKa = 4.03PRR84 pKa = 11.84RR85 pKa = 11.84GTVDD89 pKa = 3.19EE90 pKa = 4.36ARR92 pKa = 11.84TYY94 pKa = 10.65CSKK97 pKa = 10.99EE98 pKa = 3.87DD99 pKa = 3.57TRR101 pKa = 11.84EE102 pKa = 3.88PGIDD106 pKa = 3.14SGPFEE111 pKa = 5.14NGTPSKK117 pKa = 10.4SGKK120 pKa = 10.11RR121 pKa = 11.84SDD123 pKa = 3.55LADD126 pKa = 3.28IKK128 pKa = 11.22EE129 pKa = 4.06EE130 pKa = 4.05LDD132 pKa = 3.52AGSSLKK138 pKa = 10.81SISQQHH144 pKa = 6.14FGQFLRR150 pKa = 11.84YY151 pKa = 8.7RR152 pKa = 11.84KK153 pKa = 10.06SFEE156 pKa = 3.78AYY158 pKa = 8.89IVLNQDD164 pKa = 2.48PRR166 pKa = 11.84TWEE169 pKa = 3.96MEE171 pKa = 3.69NSILWGEE178 pKa = 4.2PGTGKK183 pKa = 8.67TKK185 pKa = 10.48LAYY188 pKa = 9.63DD189 pKa = 3.9LKK191 pKa = 10.99EE192 pKa = 4.03RR193 pKa = 11.84DD194 pKa = 3.72GVEE197 pKa = 4.38GYY199 pKa = 10.02PLMRR203 pKa = 11.84NQNGNVWFDD212 pKa = 3.68GYY214 pKa = 10.71HH215 pKa = 4.96GQEE218 pKa = 3.76ILLIDD223 pKa = 5.28DD224 pKa = 3.98YY225 pKa = 11.69YY226 pKa = 11.84GWIPLAFLLQLLDD239 pKa = 4.24RR240 pKa = 11.84YY241 pKa = 9.89PMNVQTKK248 pKa = 8.54GGSVPFTSKK257 pKa = 10.81KK258 pKa = 10.41IIITSNKK265 pKa = 9.87SPEE268 pKa = 3.74CWYY271 pKa = 10.48NWSKK275 pKa = 10.69FGKK278 pKa = 9.96NMFGAFEE285 pKa = 3.88RR286 pKa = 11.84RR287 pKa = 11.84INQVFHH293 pKa = 5.35YY294 pKa = 10.83VKK296 pKa = 10.78DD297 pKa = 3.82KK298 pKa = 11.43DD299 pKa = 3.52PAIYY303 pKa = 9.13PIKK306 pKa = 10.54FPEE309 pKa = 4.26TVSAVEE315 pKa = 4.46FFQQ318 pKa = 5.57
Molecular weight: 36.94 kDa
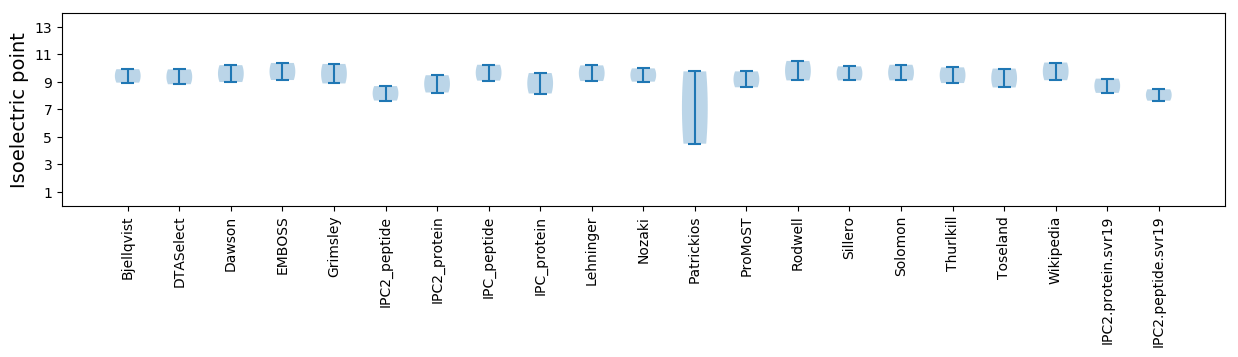
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IAX7|A0A0C5IAX7_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 7 OX=1618258 PE=4 SV=1
MM1 pKa = 6.11VTLMASFRR9 pKa = 11.84FLTFFLYY16 pKa = 10.59SLCQRR21 pKa = 11.84GNAHH25 pKa = 6.75HH26 pKa = 6.84PRR28 pKa = 11.84HH29 pKa = 6.69RR30 pKa = 11.84YY31 pKa = 9.31RR32 pKa = 11.84IIVVMGEE39 pKa = 3.83LTDD42 pKa = 3.47EE43 pKa = 4.29KK44 pKa = 10.8FRR46 pKa = 11.84IGMEE50 pKa = 3.88EE51 pKa = 3.94GLANYY56 pKa = 7.52YY57 pKa = 9.47TPQKK61 pKa = 10.43GAALVRR67 pKa = 11.84AMFPPSHH74 pKa = 5.21TWSWKK79 pKa = 7.56ARR81 pKa = 11.84KK82 pKa = 8.34WFYY85 pKa = 9.36RR86 pKa = 11.84TARR89 pKa = 11.84MRR91 pKa = 11.84MGHH94 pKa = 5.94WLYY97 pKa = 10.66LAAMFLDD104 pKa = 4.93GALTPPQVWLILRR117 pKa = 11.84SRR119 pKa = 11.84AKK121 pKa = 8.5TQQEE125 pKa = 4.18VEE127 pKa = 4.28EE128 pKa = 4.06MTEE131 pKa = 4.03AVKK134 pKa = 10.67KK135 pKa = 10.85LSLGYY140 pKa = 8.36ITKK143 pKa = 9.65IQVWCTMQKK152 pKa = 10.07RR153 pKa = 11.84EE154 pKa = 4.11VPFWMPPAWYY164 pKa = 7.9WKK166 pKa = 10.92NEE168 pKa = 3.71DD169 pKa = 3.63QYY171 pKa = 11.48WFRR174 pKa = 11.84KK175 pKa = 9.72DD176 pKa = 3.39MIWNKK181 pKa = 9.73FYY183 pKa = 11.44
MM1 pKa = 6.11VTLMASFRR9 pKa = 11.84FLTFFLYY16 pKa = 10.59SLCQRR21 pKa = 11.84GNAHH25 pKa = 6.75HH26 pKa = 6.84PRR28 pKa = 11.84HH29 pKa = 6.69RR30 pKa = 11.84YY31 pKa = 9.31RR32 pKa = 11.84IIVVMGEE39 pKa = 3.83LTDD42 pKa = 3.47EE43 pKa = 4.29KK44 pKa = 10.8FRR46 pKa = 11.84IGMEE50 pKa = 3.88EE51 pKa = 3.94GLANYY56 pKa = 7.52YY57 pKa = 9.47TPQKK61 pKa = 10.43GAALVRR67 pKa = 11.84AMFPPSHH74 pKa = 5.21TWSWKK79 pKa = 7.56ARR81 pKa = 11.84KK82 pKa = 8.34WFYY85 pKa = 9.36RR86 pKa = 11.84TARR89 pKa = 11.84MRR91 pKa = 11.84MGHH94 pKa = 5.94WLYY97 pKa = 10.66LAAMFLDD104 pKa = 4.93GALTPPQVWLILRR117 pKa = 11.84SRR119 pKa = 11.84AKK121 pKa = 8.5TQQEE125 pKa = 4.18VEE127 pKa = 4.28EE128 pKa = 4.06MTEE131 pKa = 4.03AVKK134 pKa = 10.67KK135 pKa = 10.85LSLGYY140 pKa = 8.36ITKK143 pKa = 9.65IQVWCTMQKK152 pKa = 10.07RR153 pKa = 11.84EE154 pKa = 4.11VPFWMPPAWYY164 pKa = 7.9WKK166 pKa = 10.92NEE168 pKa = 3.71DD169 pKa = 3.63QYY171 pKa = 11.48WFRR174 pKa = 11.84KK175 pKa = 9.72DD176 pKa = 3.39MIWNKK181 pKa = 9.73FYY183 pKa = 11.44
Molecular weight: 22.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
501 |
183 |
318 |
250.5 |
29.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.79 ± 1.567 | 1.198 ± 0.057 |
3.792 ± 0.88 | 6.587 ± 0.615 |
6.587 ± 0.316 | 6.587 ± 1.214 |
2.196 ± 0.294 | 5.19 ± 0.748 |
7.186 ± 0.344 | 7.585 ± 0.335 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.593 ± 1.625 | 3.393 ± 0.662 |
5.389 ± 0.258 | 4.79 ± 0.23 |
6.986 ± 0.663 | 5.389 ± 1.157 |
5.389 ± 0.341 | 4.99 ± 0.039 |
3.393 ± 1.435 | 4.99 ± 0.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
