
Halarcobacter bivalviorum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Campylobacteraceae; Arcobacter group;
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
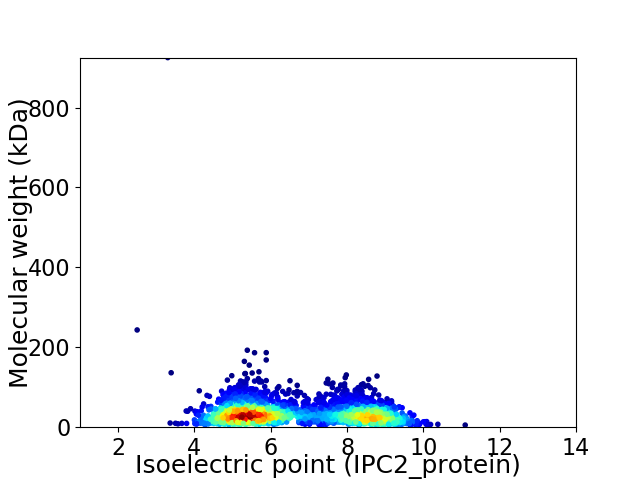
Virtual 2D-PAGE plot for 2581 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345IVM7|A0A345IVM7_9PROT NMN amidohydrolase OS=Halarcobacter bivalviorum OX=663364 GN=pncC PE=4 SV=1
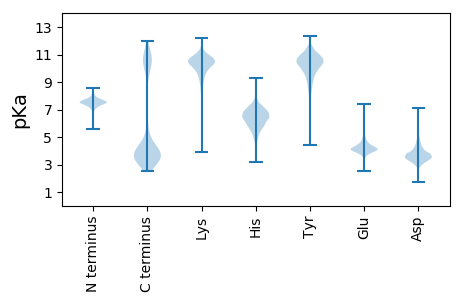
MM1 pKa = 7.61KK2 pKa = 10.26KK3 pKa = 10.6FSLFTCALLLSTSTVFAADD22 pKa = 4.16SIDD25 pKa = 3.42EE26 pKa = 4.27AFKK29 pKa = 10.74SGKK32 pKa = 9.81VSGDD36 pKa = 3.03ITLHH40 pKa = 4.52TVKK43 pKa = 10.38YY44 pKa = 9.46DD45 pKa = 3.4NKK47 pKa = 10.53GVVDD51 pKa = 3.78SGFTAGTVGLAYY63 pKa = 7.91EE64 pKa = 4.28TGSFYY69 pKa = 11.28GLSAKK74 pKa = 9.73MGFRR78 pKa = 11.84ANHH81 pKa = 5.85EE82 pKa = 4.01FSEE85 pKa = 4.54VEE87 pKa = 3.74EE88 pKa = 4.47GDD90 pKa = 3.66YY91 pKa = 11.62EE92 pKa = 4.64NFFVHH97 pKa = 7.04DD98 pKa = 4.17ALMTEE103 pKa = 5.13AYY105 pKa = 9.88LKK107 pKa = 10.36YY108 pKa = 10.33ASEE111 pKa = 4.45SFSITAGRR119 pKa = 11.84QAIDD123 pKa = 3.98LEE125 pKa = 4.2WLGDD129 pKa = 3.67YY130 pKa = 11.21NEE132 pKa = 5.59AIVAAITAIPDD143 pKa = 3.33TTIILGYY150 pKa = 7.64TDD152 pKa = 3.75RR153 pKa = 11.84QAISDD158 pKa = 4.02EE159 pKa = 4.67DD160 pKa = 3.8EE161 pKa = 4.87SSDD164 pKa = 3.64FTEE167 pKa = 4.37LTNEE171 pKa = 3.85GAYY174 pKa = 10.74VLDD177 pKa = 3.56IKK179 pKa = 11.33YY180 pKa = 10.44KK181 pKa = 10.87AFDD184 pKa = 3.46SLEE187 pKa = 4.16FNPYY191 pKa = 10.3AYY193 pKa = 10.14SAPDD197 pKa = 3.18AVDD200 pKa = 4.56FYY202 pKa = 11.69GLKK205 pKa = 9.71TSYY208 pKa = 11.09SSDD211 pKa = 2.74IFSAIAHH218 pKa = 5.86YY219 pKa = 8.07ATSNVDD225 pKa = 3.72SKK227 pKa = 11.19MKK229 pKa = 10.6DD230 pKa = 2.63ITTDD234 pKa = 3.15DD235 pKa = 3.97GNILNLEE242 pKa = 4.6LGTSFAGISASLGYY256 pKa = 10.31IKK258 pKa = 9.72TDD260 pKa = 2.8KK261 pKa = 10.98DD262 pKa = 3.6YY263 pKa = 11.76GIGLMDD269 pKa = 3.98TYY271 pKa = 11.42GDD273 pKa = 4.57NINLMDD279 pKa = 5.23SGNQIYY285 pKa = 10.43SADD288 pKa = 3.5AKK290 pKa = 9.66TVYY293 pKa = 10.6GSLSYY298 pKa = 10.59EE299 pKa = 3.8IAGVEE304 pKa = 4.02LGALYY309 pKa = 10.84SDD311 pKa = 4.12TEE313 pKa = 4.24YY314 pKa = 11.36GADD317 pKa = 3.4NFDD320 pKa = 3.72EE321 pKa = 5.36KK322 pKa = 11.12EE323 pKa = 4.08LNLTVSYY330 pKa = 10.79AITDD334 pKa = 3.86SLSVGLLYY342 pKa = 10.83ADD344 pKa = 4.84IDD346 pKa = 4.13TDD348 pKa = 5.13SDD350 pKa = 3.97DD351 pKa = 5.04ADD353 pKa = 4.93SNDD356 pKa = 3.25STYY359 pKa = 11.43GSLTLSYY366 pKa = 11.21SFF368 pKa = 4.92
MM1 pKa = 7.61KK2 pKa = 10.26KK3 pKa = 10.6FSLFTCALLLSTSTVFAADD22 pKa = 4.16SIDD25 pKa = 3.42EE26 pKa = 4.27AFKK29 pKa = 10.74SGKK32 pKa = 9.81VSGDD36 pKa = 3.03ITLHH40 pKa = 4.52TVKK43 pKa = 10.38YY44 pKa = 9.46DD45 pKa = 3.4NKK47 pKa = 10.53GVVDD51 pKa = 3.78SGFTAGTVGLAYY63 pKa = 7.91EE64 pKa = 4.28TGSFYY69 pKa = 11.28GLSAKK74 pKa = 9.73MGFRR78 pKa = 11.84ANHH81 pKa = 5.85EE82 pKa = 4.01FSEE85 pKa = 4.54VEE87 pKa = 3.74EE88 pKa = 4.47GDD90 pKa = 3.66YY91 pKa = 11.62EE92 pKa = 4.64NFFVHH97 pKa = 7.04DD98 pKa = 4.17ALMTEE103 pKa = 5.13AYY105 pKa = 9.88LKK107 pKa = 10.36YY108 pKa = 10.33ASEE111 pKa = 4.45SFSITAGRR119 pKa = 11.84QAIDD123 pKa = 3.98LEE125 pKa = 4.2WLGDD129 pKa = 3.67YY130 pKa = 11.21NEE132 pKa = 5.59AIVAAITAIPDD143 pKa = 3.33TTIILGYY150 pKa = 7.64TDD152 pKa = 3.75RR153 pKa = 11.84QAISDD158 pKa = 4.02EE159 pKa = 4.67DD160 pKa = 3.8EE161 pKa = 4.87SSDD164 pKa = 3.64FTEE167 pKa = 4.37LTNEE171 pKa = 3.85GAYY174 pKa = 10.74VLDD177 pKa = 3.56IKK179 pKa = 11.33YY180 pKa = 10.44KK181 pKa = 10.87AFDD184 pKa = 3.46SLEE187 pKa = 4.16FNPYY191 pKa = 10.3AYY193 pKa = 10.14SAPDD197 pKa = 3.18AVDD200 pKa = 4.56FYY202 pKa = 11.69GLKK205 pKa = 9.71TSYY208 pKa = 11.09SSDD211 pKa = 2.74IFSAIAHH218 pKa = 5.86YY219 pKa = 8.07ATSNVDD225 pKa = 3.72SKK227 pKa = 11.19MKK229 pKa = 10.6DD230 pKa = 2.63ITTDD234 pKa = 3.15DD235 pKa = 3.97GNILNLEE242 pKa = 4.6LGTSFAGISASLGYY256 pKa = 10.31IKK258 pKa = 9.72TDD260 pKa = 2.8KK261 pKa = 10.98DD262 pKa = 3.6YY263 pKa = 11.76GIGLMDD269 pKa = 3.98TYY271 pKa = 11.42GDD273 pKa = 4.57NINLMDD279 pKa = 5.23SGNQIYY285 pKa = 10.43SADD288 pKa = 3.5AKK290 pKa = 9.66TVYY293 pKa = 10.6GSLSYY298 pKa = 10.59EE299 pKa = 3.8IAGVEE304 pKa = 4.02LGALYY309 pKa = 10.84SDD311 pKa = 4.12TEE313 pKa = 4.24YY314 pKa = 11.36GADD317 pKa = 3.4NFDD320 pKa = 3.72EE321 pKa = 5.36KK322 pKa = 11.12EE323 pKa = 4.08LNLTVSYY330 pKa = 10.79AITDD334 pKa = 3.86SLSVGLLYY342 pKa = 10.83ADD344 pKa = 4.84IDD346 pKa = 4.13TDD348 pKa = 5.13SDD350 pKa = 3.97DD351 pKa = 5.04ADD353 pKa = 4.93SNDD356 pKa = 3.25STYY359 pKa = 11.43GSLTLSYY366 pKa = 11.21SFF368 pKa = 4.92
Molecular weight: 39.95 kDa
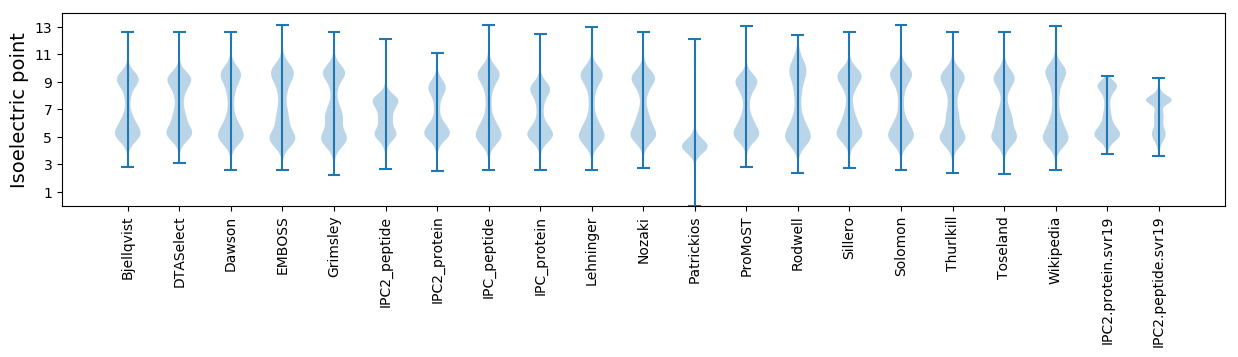
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345IZL4|A0A345IZL4_9PROT DUF59 domain-containing protein OS=Halarcobacter bivalviorum OX=663364 GN=ABIV_2021 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.31RR14 pKa = 11.84THH16 pKa = 6.08GFRR19 pKa = 11.84IRR21 pKa = 11.84MKK23 pKa = 9.04TKK25 pKa = 10.16NGRR28 pKa = 11.84KK29 pKa = 8.88VIKK32 pKa = 10.14ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.31RR14 pKa = 11.84THH16 pKa = 6.08GFRR19 pKa = 11.84IRR21 pKa = 11.84MKK23 pKa = 9.04TKK25 pKa = 10.16NGRR28 pKa = 11.84KK29 pKa = 8.88VIKK32 pKa = 10.14ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
831417 |
37 |
8997 |
322.1 |
36.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.909 ± 0.053 | 0.861 ± 0.02 |
5.682 ± 0.128 | 7.649 ± 0.067 |
5.267 ± 0.059 | 5.311 ± 0.114 |
1.561 ± 0.022 | 9.351 ± 0.057 |
9.587 ± 0.101 | 9.836 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.231 ± 0.034 | 6.197 ± 0.068 |
2.619 ± 0.037 | 2.758 ± 0.026 |
2.901 ± 0.034 | 6.557 ± 0.047 |
5.171 ± 0.095 | 5.854 ± 0.038 |
0.677 ± 0.017 | 4.02 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
