
Melon partitivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 5.74
Get precalculated fractions of proteins
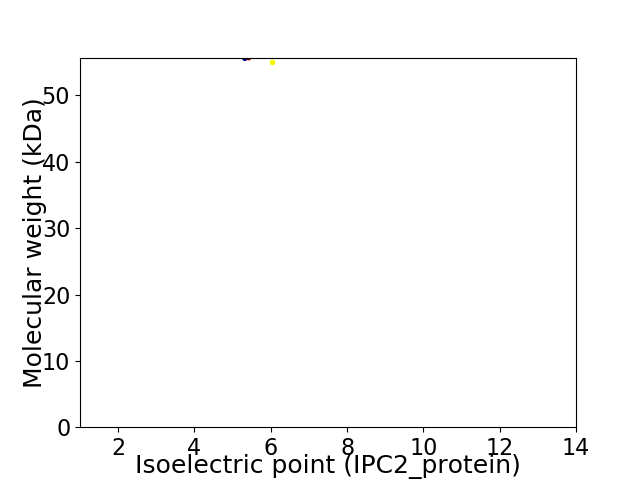
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346RRG4|A0A346RRG4_9VIRU Putative coat protein OS=Melon partitivirus OX=2305257 GN=CP PE=4 SV=1
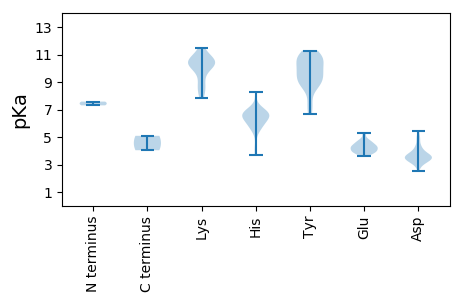
MM1 pKa = 7.58AALGDD6 pKa = 4.39GVPPIADD13 pKa = 3.35NNNPQGRR20 pKa = 11.84DD21 pKa = 3.25ANPIRR26 pKa = 11.84GGTDD30 pKa = 3.19PLRR33 pKa = 11.84RR34 pKa = 11.84ATDD37 pKa = 3.27WLEE40 pKa = 4.42DD41 pKa = 3.47NSKK44 pKa = 8.63ITMTFHH50 pKa = 6.76RR51 pKa = 11.84HH52 pKa = 3.67ANYY55 pKa = 9.86RR56 pKa = 11.84RR57 pKa = 11.84YY58 pKa = 9.1ATLRR62 pKa = 11.84LQEE65 pKa = 4.2MFNALVQLYY74 pKa = 9.5QNIFMSHH81 pKa = 4.26WHH83 pKa = 5.84HH84 pKa = 7.02LRR86 pKa = 11.84RR87 pKa = 11.84TVEE90 pKa = 4.09NYY92 pKa = 8.94QVPAPLAQTPWLYY105 pKa = 10.66VARR108 pKa = 11.84LYY110 pKa = 10.67ISVWIVDD117 pKa = 4.38LYY119 pKa = 11.28ASIRR123 pKa = 11.84RR124 pKa = 11.84ACLRR128 pKa = 11.84VCPEE132 pKa = 4.62AYY134 pKa = 10.08NEE136 pKa = 4.77YY137 pKa = 9.11YY138 pKa = 8.62TQDD141 pKa = 3.35YY142 pKa = 8.56QQVVFEE148 pKa = 4.31YY149 pKa = 11.01DD150 pKa = 3.0AFLSQLLSMIKK161 pKa = 8.46PTVVNLVHH169 pKa = 6.21EE170 pKa = 4.66TTLFVPRR177 pKa = 11.84LAEE180 pKa = 4.29DD181 pKa = 3.65QPFPADD187 pKa = 3.57DD188 pKa = 3.7MNIFGLDD195 pKa = 2.81RR196 pKa = 11.84FDD198 pKa = 4.15YY199 pKa = 11.0NEE201 pKa = 4.03NVVHH205 pKa = 6.37GVISILRR212 pKa = 11.84DD213 pKa = 3.21RR214 pKa = 11.84KK215 pKa = 10.4LMKK218 pKa = 10.17FEE220 pKa = 4.84KK221 pKa = 10.03PSEE224 pKa = 4.27TGFGRR229 pKa = 11.84PFWLFDD235 pKa = 2.98WHH237 pKa = 6.15RR238 pKa = 11.84TGHH241 pKa = 6.24AYY243 pKa = 10.23AWFPAEE249 pKa = 4.07NNYY252 pKa = 10.82DD253 pKa = 3.59MRR255 pKa = 11.84DD256 pKa = 3.3VTVAYY261 pKa = 9.37IVGVACTPKK270 pKa = 10.53LGPRR274 pKa = 11.84DD275 pKa = 3.4IDD277 pKa = 3.55DD278 pKa = 3.7WQHH281 pKa = 5.74FPGNIIPEE289 pKa = 4.09HH290 pKa = 6.55PRR292 pKa = 11.84AEE294 pKa = 3.94DD295 pKa = 3.34HH296 pKa = 6.61VRR298 pKa = 11.84VRR300 pKa = 11.84PVAYY304 pKa = 9.75HH305 pKa = 5.88GAVEE309 pKa = 4.11VRR311 pKa = 11.84TIDD314 pKa = 3.62VQQWTAPPEE323 pKa = 4.25VMSLIQVTLPPAAAQQQQQQQPAAQAQPAPARR355 pKa = 11.84QQPPAQQQQQEE366 pKa = 4.23QGLIRR371 pKa = 11.84YY372 pKa = 7.35NLRR375 pKa = 11.84QGFKK379 pKa = 10.33RR380 pKa = 11.84RR381 pKa = 11.84RR382 pKa = 11.84TDD384 pKa = 3.04PSTSNDD390 pKa = 2.52NDD392 pKa = 3.66MPVEE396 pKa = 4.79DD397 pKa = 4.77IEE399 pKa = 5.26EE400 pKa = 4.53PPQPEE405 pKa = 4.01QGEE408 pKa = 4.47DD409 pKa = 3.67PIHH412 pKa = 6.62PEE414 pKa = 3.86EE415 pKa = 4.34EE416 pKa = 3.79AAPPVEE422 pKa = 4.51PQPDD426 pKa = 4.06PAPAPAPAPQAPAEE440 pKa = 4.59PIITMNRR447 pKa = 11.84VRR449 pKa = 11.84RR450 pKa = 11.84FRR452 pKa = 11.84IITYY456 pKa = 9.39CYY458 pKa = 9.13YY459 pKa = 10.71RR460 pKa = 11.84QVTDD464 pKa = 4.34NEE466 pKa = 4.45DD467 pKa = 3.17LHH469 pKa = 8.3SRR471 pKa = 11.84LAALRR476 pKa = 11.84QVDD479 pKa = 3.92SLIAA483 pKa = 4.04
MM1 pKa = 7.58AALGDD6 pKa = 4.39GVPPIADD13 pKa = 3.35NNNPQGRR20 pKa = 11.84DD21 pKa = 3.25ANPIRR26 pKa = 11.84GGTDD30 pKa = 3.19PLRR33 pKa = 11.84RR34 pKa = 11.84ATDD37 pKa = 3.27WLEE40 pKa = 4.42DD41 pKa = 3.47NSKK44 pKa = 8.63ITMTFHH50 pKa = 6.76RR51 pKa = 11.84HH52 pKa = 3.67ANYY55 pKa = 9.86RR56 pKa = 11.84RR57 pKa = 11.84YY58 pKa = 9.1ATLRR62 pKa = 11.84LQEE65 pKa = 4.2MFNALVQLYY74 pKa = 9.5QNIFMSHH81 pKa = 4.26WHH83 pKa = 5.84HH84 pKa = 7.02LRR86 pKa = 11.84RR87 pKa = 11.84TVEE90 pKa = 4.09NYY92 pKa = 8.94QVPAPLAQTPWLYY105 pKa = 10.66VARR108 pKa = 11.84LYY110 pKa = 10.67ISVWIVDD117 pKa = 4.38LYY119 pKa = 11.28ASIRR123 pKa = 11.84RR124 pKa = 11.84ACLRR128 pKa = 11.84VCPEE132 pKa = 4.62AYY134 pKa = 10.08NEE136 pKa = 4.77YY137 pKa = 9.11YY138 pKa = 8.62TQDD141 pKa = 3.35YY142 pKa = 8.56QQVVFEE148 pKa = 4.31YY149 pKa = 11.01DD150 pKa = 3.0AFLSQLLSMIKK161 pKa = 8.46PTVVNLVHH169 pKa = 6.21EE170 pKa = 4.66TTLFVPRR177 pKa = 11.84LAEE180 pKa = 4.29DD181 pKa = 3.65QPFPADD187 pKa = 3.57DD188 pKa = 3.7MNIFGLDD195 pKa = 2.81RR196 pKa = 11.84FDD198 pKa = 4.15YY199 pKa = 11.0NEE201 pKa = 4.03NVVHH205 pKa = 6.37GVISILRR212 pKa = 11.84DD213 pKa = 3.21RR214 pKa = 11.84KK215 pKa = 10.4LMKK218 pKa = 10.17FEE220 pKa = 4.84KK221 pKa = 10.03PSEE224 pKa = 4.27TGFGRR229 pKa = 11.84PFWLFDD235 pKa = 2.98WHH237 pKa = 6.15RR238 pKa = 11.84TGHH241 pKa = 6.24AYY243 pKa = 10.23AWFPAEE249 pKa = 4.07NNYY252 pKa = 10.82DD253 pKa = 3.59MRR255 pKa = 11.84DD256 pKa = 3.3VTVAYY261 pKa = 9.37IVGVACTPKK270 pKa = 10.53LGPRR274 pKa = 11.84DD275 pKa = 3.4IDD277 pKa = 3.55DD278 pKa = 3.7WQHH281 pKa = 5.74FPGNIIPEE289 pKa = 4.09HH290 pKa = 6.55PRR292 pKa = 11.84AEE294 pKa = 3.94DD295 pKa = 3.34HH296 pKa = 6.61VRR298 pKa = 11.84VRR300 pKa = 11.84PVAYY304 pKa = 9.75HH305 pKa = 5.88GAVEE309 pKa = 4.11VRR311 pKa = 11.84TIDD314 pKa = 3.62VQQWTAPPEE323 pKa = 4.25VMSLIQVTLPPAAAQQQQQQQPAAQAQPAPARR355 pKa = 11.84QQPPAQQQQQEE366 pKa = 4.23QGLIRR371 pKa = 11.84YY372 pKa = 7.35NLRR375 pKa = 11.84QGFKK379 pKa = 10.33RR380 pKa = 11.84RR381 pKa = 11.84RR382 pKa = 11.84TDD384 pKa = 3.04PSTSNDD390 pKa = 2.52NDD392 pKa = 3.66MPVEE396 pKa = 4.79DD397 pKa = 4.77IEE399 pKa = 5.26EE400 pKa = 4.53PPQPEE405 pKa = 4.01QGEE408 pKa = 4.47DD409 pKa = 3.67PIHH412 pKa = 6.62PEE414 pKa = 3.86EE415 pKa = 4.34EE416 pKa = 3.79AAPPVEE422 pKa = 4.51PQPDD426 pKa = 4.06PAPAPAPAPQAPAEE440 pKa = 4.59PIITMNRR447 pKa = 11.84VRR449 pKa = 11.84RR450 pKa = 11.84FRR452 pKa = 11.84IITYY456 pKa = 9.39CYY458 pKa = 9.13YY459 pKa = 10.71RR460 pKa = 11.84QVTDD464 pKa = 4.34NEE466 pKa = 4.45DD467 pKa = 3.17LHH469 pKa = 8.3SRR471 pKa = 11.84LAALRR476 pKa = 11.84QVDD479 pKa = 3.92SLIAA483 pKa = 4.04
Molecular weight: 55.61 kDa
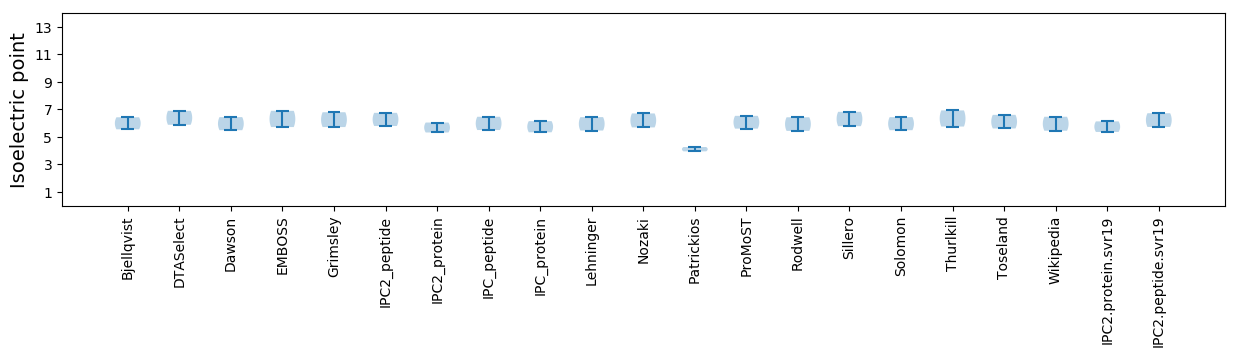
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346RRG4|A0A346RRG4_9VIRU Putative coat protein OS=Melon partitivirus OX=2305257 GN=CP PE=4 SV=1
MM1 pKa = 7.3LRR3 pKa = 11.84DD4 pKa = 3.32IRR6 pKa = 11.84NFQLTEE12 pKa = 4.09FNSEE16 pKa = 4.17FEE18 pKa = 4.73LLNDD22 pKa = 3.68THH24 pKa = 6.32PHH26 pKa = 5.6VIRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 4.03AEE33 pKa = 3.85QILVDD38 pKa = 4.1EE39 pKa = 5.0FALSEE44 pKa = 4.8LNGAWPSLYY53 pKa = 10.09DD54 pKa = 3.51QKK56 pKa = 11.5LKK58 pKa = 10.36GWARR62 pKa = 11.84SFYY65 pKa = 9.74TLEE68 pKa = 3.64GHH70 pKa = 5.79MQAIHH75 pKa = 7.02AYY77 pKa = 9.37SNPDD81 pKa = 3.38TPISSLNEE89 pKa = 3.69TIYY92 pKa = 10.8RR93 pKa = 11.84ATIDD97 pKa = 3.43SLKK100 pKa = 10.94NEE102 pKa = 4.31LSSLPTARR110 pKa = 11.84AFDD113 pKa = 3.8VLTEE117 pKa = 3.99LDD119 pKa = 4.08KK120 pKa = 11.15IHH122 pKa = 6.54YY123 pKa = 6.93EE124 pKa = 3.87QSSAAGYY131 pKa = 10.74NYY133 pKa = 9.8IGPKK137 pKa = 9.72GPLYY141 pKa = 10.92GEE143 pKa = 3.8NHH145 pKa = 5.44EE146 pKa = 4.23RR147 pKa = 11.84AIRR150 pKa = 11.84SARR153 pKa = 11.84AVLWSAIRR161 pKa = 11.84DD162 pKa = 3.77EE163 pKa = 5.27DD164 pKa = 3.76GGPAYY169 pKa = 10.4VLRR172 pKa = 11.84NMVPDD177 pKa = 3.51VGYY180 pKa = 8.76TRR182 pKa = 11.84TQLTNLQEE190 pKa = 4.12KK191 pKa = 8.94TKK193 pKa = 10.58VRR195 pKa = 11.84GVWGRR200 pKa = 11.84AFHH203 pKa = 6.66YY204 pKa = 10.28ILLEE208 pKa = 4.32GSSARR213 pKa = 11.84PLLEE217 pKa = 4.37RR218 pKa = 11.84FLEE221 pKa = 4.51HH222 pKa = 6.64DD223 pKa = 3.56TFYY226 pKa = 10.99HH227 pKa = 6.82IGEE230 pKa = 4.65DD231 pKa = 3.71SQLSVPKK238 pKa = 10.54LMSKK242 pKa = 10.59LSSEE246 pKa = 4.45CRR248 pKa = 11.84WIYY251 pKa = 10.84AIDD254 pKa = 3.54WSAFDD259 pKa = 3.51ATVSRR264 pKa = 11.84FEE266 pKa = 3.87IEE268 pKa = 3.75AAFEE272 pKa = 3.98IMKK275 pKa = 10.64GLIHH279 pKa = 6.79FPNFDD284 pKa = 3.53SEE286 pKa = 4.65QAFEE290 pKa = 4.2LSRR293 pKa = 11.84LLFTHH298 pKa = 6.76KK299 pKa = 10.38KK300 pKa = 8.26IAAPDD305 pKa = 3.27GHH307 pKa = 6.56TYY309 pKa = 9.86WIHH312 pKa = 6.71KK313 pKa = 7.85GIPSGSYY320 pKa = 6.7FTSMAGSIINRR331 pKa = 11.84LRR333 pKa = 11.84IGYY336 pKa = 8.8LWRR339 pKa = 11.84LMFNRR344 pKa = 11.84EE345 pKa = 3.67PKK347 pKa = 10.31LCYY350 pKa = 9.16TQGDD354 pKa = 3.91DD355 pKa = 5.12SLIGDD360 pKa = 4.86DD361 pKa = 5.46EE362 pKa = 5.31LIQPEE367 pKa = 4.37RR368 pKa = 11.84LAEE371 pKa = 3.93LAAPLNWNINPNKK384 pKa = 9.27TEE386 pKa = 3.87YY387 pKa = 10.84SRR389 pKa = 11.84TPEE392 pKa = 4.34FISFLGRR399 pKa = 11.84TSRR402 pKa = 11.84GGFNTRR408 pKa = 11.84EE409 pKa = 3.96LEE411 pKa = 3.86KK412 pKa = 10.69CLRR415 pKa = 11.84LLIFPEE421 pKa = 4.32HH422 pKa = 6.71PVPSGRR428 pKa = 11.84ISAYY432 pKa = 8.99RR433 pKa = 11.84ARR435 pKa = 11.84SINEE439 pKa = 3.68DD440 pKa = 3.39SGFTSDD446 pKa = 4.99YY447 pKa = 8.73IQTVARR453 pKa = 11.84RR454 pKa = 11.84LRR456 pKa = 11.84RR457 pKa = 11.84RR458 pKa = 11.84YY459 pKa = 9.99GIAEE463 pKa = 3.98EE464 pKa = 4.61SEE466 pKa = 4.24VPKK469 pKa = 10.19MFKK472 pKa = 10.21RR473 pKa = 11.84YY474 pKa = 7.63VVRR477 pKa = 5.09
MM1 pKa = 7.3LRR3 pKa = 11.84DD4 pKa = 3.32IRR6 pKa = 11.84NFQLTEE12 pKa = 4.09FNSEE16 pKa = 4.17FEE18 pKa = 4.73LLNDD22 pKa = 3.68THH24 pKa = 6.32PHH26 pKa = 5.6VIRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 4.03AEE33 pKa = 3.85QILVDD38 pKa = 4.1EE39 pKa = 5.0FALSEE44 pKa = 4.8LNGAWPSLYY53 pKa = 10.09DD54 pKa = 3.51QKK56 pKa = 11.5LKK58 pKa = 10.36GWARR62 pKa = 11.84SFYY65 pKa = 9.74TLEE68 pKa = 3.64GHH70 pKa = 5.79MQAIHH75 pKa = 7.02AYY77 pKa = 9.37SNPDD81 pKa = 3.38TPISSLNEE89 pKa = 3.69TIYY92 pKa = 10.8RR93 pKa = 11.84ATIDD97 pKa = 3.43SLKK100 pKa = 10.94NEE102 pKa = 4.31LSSLPTARR110 pKa = 11.84AFDD113 pKa = 3.8VLTEE117 pKa = 3.99LDD119 pKa = 4.08KK120 pKa = 11.15IHH122 pKa = 6.54YY123 pKa = 6.93EE124 pKa = 3.87QSSAAGYY131 pKa = 10.74NYY133 pKa = 9.8IGPKK137 pKa = 9.72GPLYY141 pKa = 10.92GEE143 pKa = 3.8NHH145 pKa = 5.44EE146 pKa = 4.23RR147 pKa = 11.84AIRR150 pKa = 11.84SARR153 pKa = 11.84AVLWSAIRR161 pKa = 11.84DD162 pKa = 3.77EE163 pKa = 5.27DD164 pKa = 3.76GGPAYY169 pKa = 10.4VLRR172 pKa = 11.84NMVPDD177 pKa = 3.51VGYY180 pKa = 8.76TRR182 pKa = 11.84TQLTNLQEE190 pKa = 4.12KK191 pKa = 8.94TKK193 pKa = 10.58VRR195 pKa = 11.84GVWGRR200 pKa = 11.84AFHH203 pKa = 6.66YY204 pKa = 10.28ILLEE208 pKa = 4.32GSSARR213 pKa = 11.84PLLEE217 pKa = 4.37RR218 pKa = 11.84FLEE221 pKa = 4.51HH222 pKa = 6.64DD223 pKa = 3.56TFYY226 pKa = 10.99HH227 pKa = 6.82IGEE230 pKa = 4.65DD231 pKa = 3.71SQLSVPKK238 pKa = 10.54LMSKK242 pKa = 10.59LSSEE246 pKa = 4.45CRR248 pKa = 11.84WIYY251 pKa = 10.84AIDD254 pKa = 3.54WSAFDD259 pKa = 3.51ATVSRR264 pKa = 11.84FEE266 pKa = 3.87IEE268 pKa = 3.75AAFEE272 pKa = 3.98IMKK275 pKa = 10.64GLIHH279 pKa = 6.79FPNFDD284 pKa = 3.53SEE286 pKa = 4.65QAFEE290 pKa = 4.2LSRR293 pKa = 11.84LLFTHH298 pKa = 6.76KK299 pKa = 10.38KK300 pKa = 8.26IAAPDD305 pKa = 3.27GHH307 pKa = 6.56TYY309 pKa = 9.86WIHH312 pKa = 6.71KK313 pKa = 7.85GIPSGSYY320 pKa = 6.7FTSMAGSIINRR331 pKa = 11.84LRR333 pKa = 11.84IGYY336 pKa = 8.8LWRR339 pKa = 11.84LMFNRR344 pKa = 11.84EE345 pKa = 3.67PKK347 pKa = 10.31LCYY350 pKa = 9.16TQGDD354 pKa = 3.91DD355 pKa = 5.12SLIGDD360 pKa = 4.86DD361 pKa = 5.46EE362 pKa = 5.31LIQPEE367 pKa = 4.37RR368 pKa = 11.84LAEE371 pKa = 3.93LAAPLNWNINPNKK384 pKa = 9.27TEE386 pKa = 3.87YY387 pKa = 10.84SRR389 pKa = 11.84TPEE392 pKa = 4.34FISFLGRR399 pKa = 11.84TSRR402 pKa = 11.84GGFNTRR408 pKa = 11.84EE409 pKa = 3.96LEE411 pKa = 3.86KK412 pKa = 10.69CLRR415 pKa = 11.84LLIFPEE421 pKa = 4.32HH422 pKa = 6.71PVPSGRR428 pKa = 11.84ISAYY432 pKa = 8.99RR433 pKa = 11.84ARR435 pKa = 11.84SINEE439 pKa = 3.68DD440 pKa = 3.39SGFTSDD446 pKa = 4.99YY447 pKa = 8.73IQTVARR453 pKa = 11.84RR454 pKa = 11.84LRR456 pKa = 11.84RR457 pKa = 11.84RR458 pKa = 11.84YY459 pKa = 9.99GIAEE463 pKa = 3.98EE464 pKa = 4.61SEE466 pKa = 4.24VPKK469 pKa = 10.19MFKK472 pKa = 10.21RR473 pKa = 11.84YY474 pKa = 7.63VVRR477 pKa = 5.09
Molecular weight: 54.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
960 |
477 |
483 |
480.0 |
55.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.333 ± 0.789 | 0.729 ± 0.066 |
5.729 ± 0.594 | 6.979 ± 0.784 |
4.271 ± 0.498 | 4.688 ± 0.774 |
3.021 ± 0.056 | 6.042 ± 0.574 |
2.604 ± 0.766 | 8.333 ± 1.133 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.979 ± 0.198 | 4.375 ± 0.119 |
7.5 ± 1.754 | 5.313 ± 1.831 |
8.229 ± 0.103 | 5.417 ± 1.807 |
4.896 ± 0.089 | 5.208 ± 1.214 |
1.875 ± 0.008 | 4.479 ± 0.087 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
