
Treponema succinifaciens (strain ATCC 33096 / DSM 2489 / 6091)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Treponemataceae; Treponema; Treponema succinifaciens
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
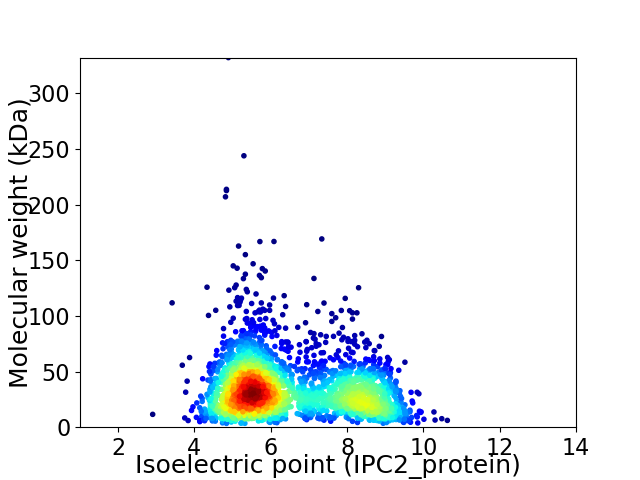
Virtual 2D-PAGE plot for 2561 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2NS82|F2NS82_TRES6 Chaperone protein ClpB OS=Treponema succinifaciens (strain ATCC 33096 / DSM 2489 / 6091) OX=869209 GN=clpB PE=3 SV=1
MM1 pKa = 6.99QFFWGGALISCSNGSSSSDD20 pKa = 3.35EE21 pKa = 5.0DD22 pKa = 4.58DD23 pKa = 5.47DD24 pKa = 4.39EE25 pKa = 6.04SSYY28 pKa = 9.57GTKK31 pKa = 9.53IYY33 pKa = 10.64SEE35 pKa = 4.42SVTFSEE41 pKa = 4.95TPTQIVSAEE50 pKa = 4.08TLADD54 pKa = 4.04LSIKK58 pKa = 10.62SLTIKK63 pKa = 10.23VINVNYY69 pKa = 10.27SGSDD73 pKa = 3.37DD74 pKa = 3.14WWFTACSDD82 pKa = 4.1SSWSNEE88 pKa = 4.29AKK90 pKa = 10.82LEE92 pKa = 4.09WQSGEE97 pKa = 4.17GTNSLYY103 pKa = 10.95LATITDD109 pKa = 3.9AAAIAAYY116 pKa = 8.61KK117 pKa = 10.86ANGIYY122 pKa = 10.03IAGTDD127 pKa = 3.41SATAAVSVFYY137 pKa = 11.39VEE139 pKa = 5.73GEE141 pKa = 4.12AEE143 pKa = 4.04EE144 pKa = 5.02SNASSGSDD152 pKa = 2.99SQTEE156 pKa = 4.01NTTTSNEE163 pKa = 4.18TSSSEE168 pKa = 4.05NEE170 pKa = 4.13TTGTSSNEE178 pKa = 3.79NEE180 pKa = 4.54DD181 pKa = 3.73NGSSSEE187 pKa = 3.98EE188 pKa = 3.61ALIEE192 pKa = 4.28SFSSNTRR199 pKa = 11.84SANFTISSSYY209 pKa = 9.38QLLMKK214 pKa = 10.8YY215 pKa = 7.55EE216 pKa = 4.45TVNSASKK223 pKa = 9.12VTITISDD230 pKa = 3.87VQTGTSNGDD239 pKa = 2.08WWFASGTDD247 pKa = 3.58SSKK250 pKa = 10.44WKK252 pKa = 10.66ADD254 pKa = 3.51IAWKK258 pKa = 9.87EE259 pKa = 3.75GNYY262 pKa = 9.98YY263 pKa = 9.99EE264 pKa = 5.95LEE266 pKa = 3.98ISDD269 pKa = 3.07ISEE272 pKa = 4.1YY273 pKa = 11.27SKK275 pKa = 11.15GIYY278 pKa = 10.02LGGLEE283 pKa = 4.37GLSGTVTVSYY293 pKa = 10.32EE294 pKa = 3.68
MM1 pKa = 6.99QFFWGGALISCSNGSSSSDD20 pKa = 3.35EE21 pKa = 5.0DD22 pKa = 4.58DD23 pKa = 5.47DD24 pKa = 4.39EE25 pKa = 6.04SSYY28 pKa = 9.57GTKK31 pKa = 9.53IYY33 pKa = 10.64SEE35 pKa = 4.42SVTFSEE41 pKa = 4.95TPTQIVSAEE50 pKa = 4.08TLADD54 pKa = 4.04LSIKK58 pKa = 10.62SLTIKK63 pKa = 10.23VINVNYY69 pKa = 10.27SGSDD73 pKa = 3.37DD74 pKa = 3.14WWFTACSDD82 pKa = 4.1SSWSNEE88 pKa = 4.29AKK90 pKa = 10.82LEE92 pKa = 4.09WQSGEE97 pKa = 4.17GTNSLYY103 pKa = 10.95LATITDD109 pKa = 3.9AAAIAAYY116 pKa = 8.61KK117 pKa = 10.86ANGIYY122 pKa = 10.03IAGTDD127 pKa = 3.41SATAAVSVFYY137 pKa = 11.39VEE139 pKa = 5.73GEE141 pKa = 4.12AEE143 pKa = 4.04EE144 pKa = 5.02SNASSGSDD152 pKa = 2.99SQTEE156 pKa = 4.01NTTTSNEE163 pKa = 4.18TSSSEE168 pKa = 4.05NEE170 pKa = 4.13TTGTSSNEE178 pKa = 3.79NEE180 pKa = 4.54DD181 pKa = 3.73NGSSSEE187 pKa = 3.98EE188 pKa = 3.61ALIEE192 pKa = 4.28SFSSNTRR199 pKa = 11.84SANFTISSSYY209 pKa = 9.38QLLMKK214 pKa = 10.8YY215 pKa = 7.55EE216 pKa = 4.45TVNSASKK223 pKa = 9.12VTITISDD230 pKa = 3.87VQTGTSNGDD239 pKa = 2.08WWFASGTDD247 pKa = 3.58SSKK250 pKa = 10.44WKK252 pKa = 10.66ADD254 pKa = 3.51IAWKK258 pKa = 9.87EE259 pKa = 3.75GNYY262 pKa = 9.98YY263 pKa = 9.99EE264 pKa = 5.95LEE266 pKa = 3.98ISDD269 pKa = 3.07ISEE272 pKa = 4.1YY273 pKa = 11.27SKK275 pKa = 11.15GIYY278 pKa = 10.02LGGLEE283 pKa = 4.37GLSGTVTVSYY293 pKa = 10.32EE294 pKa = 3.68
Molecular weight: 31.45 kDa
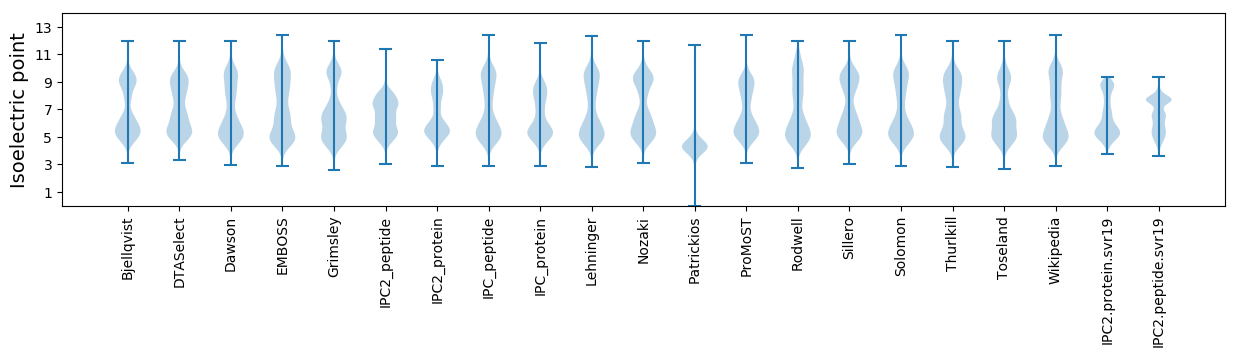
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2NSN6|F2NSN6_TRES6 Uncharacterized protein OS=Treponema succinifaciens (strain ATCC 33096 / DSM 2489 / 6091) OX=869209 GN=Tresu_1927 PE=4 SV=1
MM1 pKa = 7.67VLISVFLYY9 pKa = 8.43TVRR12 pKa = 11.84FPYY15 pKa = 10.48FKK17 pKa = 10.57RR18 pKa = 11.84IDD20 pKa = 3.41FRR22 pKa = 11.84TRR24 pKa = 11.84QVRR27 pKa = 11.84FSALRR32 pKa = 11.84RR33 pKa = 11.84TITGFLEE40 pKa = 3.87VGVWNRR46 pKa = 11.84FIGFRR51 pKa = 11.84VNAVGFSTALFLDD64 pKa = 4.15FHH66 pKa = 6.65QPRR69 pKa = 11.84ILQLPQGVDD78 pKa = 2.87RR79 pKa = 11.84FLPPAAEE86 pKa = 4.21QSDD89 pKa = 3.86HH90 pKa = 6.77LADD93 pKa = 3.77GVVQVDD99 pKa = 3.8PPVLVRR105 pKa = 11.84PAVLPGQLRR114 pKa = 11.84PAQDD118 pKa = 3.12KK119 pKa = 10.94GIQHH123 pKa = 6.71LRR125 pKa = 11.84FVAQGLEE132 pKa = 4.13CGCFKK137 pKa = 11.06KK138 pKa = 10.75EE139 pKa = 3.51IGEE142 pKa = 4.28PGEE145 pKa = 4.11TEE147 pKa = 3.68RR148 pKa = 11.84LFRR151 pKa = 11.84LVNINRR157 pKa = 11.84VCHH160 pKa = 6.16SVFCGRR166 pKa = 11.84LEE168 pKa = 3.77ARR170 pKa = 11.84FPGRR174 pKa = 11.84MINDD178 pKa = 3.75TSRR181 pKa = 11.84PGG183 pKa = 3.06
MM1 pKa = 7.67VLISVFLYY9 pKa = 8.43TVRR12 pKa = 11.84FPYY15 pKa = 10.48FKK17 pKa = 10.57RR18 pKa = 11.84IDD20 pKa = 3.41FRR22 pKa = 11.84TRR24 pKa = 11.84QVRR27 pKa = 11.84FSALRR32 pKa = 11.84RR33 pKa = 11.84TITGFLEE40 pKa = 3.87VGVWNRR46 pKa = 11.84FIGFRR51 pKa = 11.84VNAVGFSTALFLDD64 pKa = 4.15FHH66 pKa = 6.65QPRR69 pKa = 11.84ILQLPQGVDD78 pKa = 2.87RR79 pKa = 11.84FLPPAAEE86 pKa = 4.21QSDD89 pKa = 3.86HH90 pKa = 6.77LADD93 pKa = 3.77GVVQVDD99 pKa = 3.8PPVLVRR105 pKa = 11.84PAVLPGQLRR114 pKa = 11.84PAQDD118 pKa = 3.12KK119 pKa = 10.94GIQHH123 pKa = 6.71LRR125 pKa = 11.84FVAQGLEE132 pKa = 4.13CGCFKK137 pKa = 11.06KK138 pKa = 10.75EE139 pKa = 3.51IGEE142 pKa = 4.28PGEE145 pKa = 4.11TEE147 pKa = 3.68RR148 pKa = 11.84LFRR151 pKa = 11.84LVNINRR157 pKa = 11.84VCHH160 pKa = 6.16SVFCGRR166 pKa = 11.84LEE168 pKa = 3.77ARR170 pKa = 11.84FPGRR174 pKa = 11.84MINDD178 pKa = 3.75TSRR181 pKa = 11.84PGG183 pKa = 3.06
Molecular weight: 20.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
829679 |
30 |
2901 |
324.0 |
36.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.95 ± 0.043 | 1.49 ± 0.02 |
5.367 ± 0.041 | 7.477 ± 0.055 |
5.371 ± 0.046 | 6.199 ± 0.046 |
1.44 ± 0.017 | 7.66 ± 0.039 |
8.436 ± 0.046 | 8.72 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.257 ± 0.02 | 5.396 ± 0.04 |
3.322 ± 0.028 | 2.991 ± 0.024 |
3.765 ± 0.036 | 7.441 ± 0.048 |
4.958 ± 0.036 | 6.111 ± 0.045 |
0.954 ± 0.016 | 3.692 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
