
Equine foamy virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Equispumavirus
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
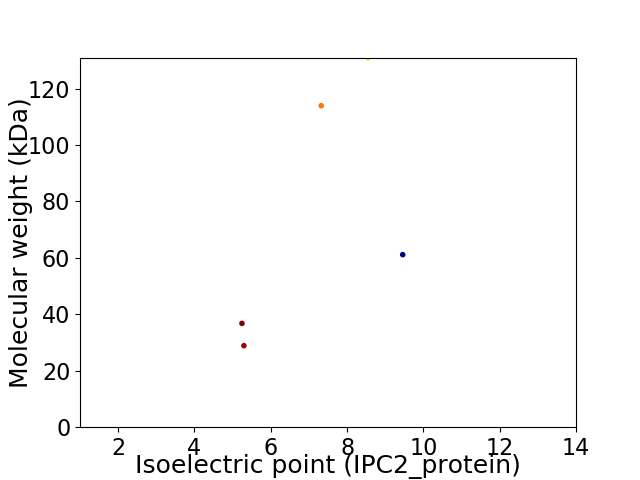
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
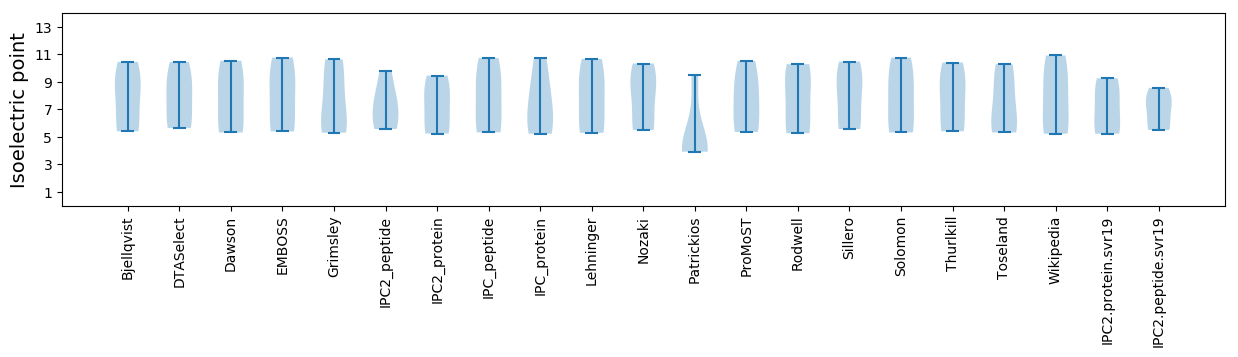
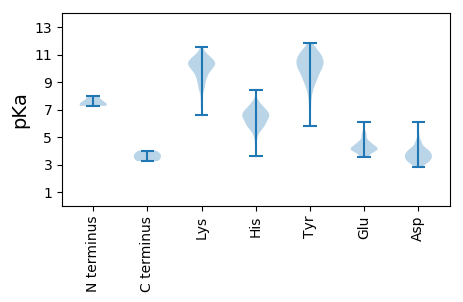
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9J4C5|Q9J4C5_9RETR ORF1/Tas/Bel1 OS=Equine foamy virus OX=109270 GN=tas PE=4 SV=1
MM1 pKa = 8.0RR2 pKa = 11.84NPGLGCLTWTGQLNGMGRR20 pKa = 11.84FQIQLFRR27 pKa = 11.84NLTLISVCLGSDD39 pKa = 3.19HH40 pKa = 7.15HH41 pKa = 8.38KK42 pKa = 11.12GITDD46 pKa = 3.65YY47 pKa = 11.41CCGPASCYY55 pKa = 9.72TIVWEE60 pKa = 4.65ISAEE64 pKa = 4.11VLATGTPYY72 pKa = 11.03LFRR75 pKa = 11.84AQPHH79 pKa = 5.94HH80 pKa = 7.2RR81 pKa = 11.84PLQTPAPWSKK91 pKa = 10.32RR92 pKa = 11.84LIRR95 pKa = 11.84TRR97 pKa = 11.84YY98 pKa = 8.45IRR100 pKa = 11.84NSVEE104 pKa = 3.66DD105 pKa = 3.71QFVHH109 pKa = 6.91FATEE113 pKa = 3.69ANYY116 pKa = 9.91TPLSAQALSIKK127 pKa = 10.11AIRR130 pKa = 11.84WHH132 pKa = 6.6KK133 pKa = 10.28GQLACEE139 pKa = 4.07GTPRR143 pKa = 11.84IGQAGIPEE151 pKa = 4.72TIYY154 pKa = 11.11AGIKK158 pKa = 9.37AYY160 pKa = 10.19GWFIPGKK167 pKa = 9.21KK168 pKa = 9.52EE169 pKa = 3.56RR170 pKa = 11.84VSWNEE175 pKa = 3.53DD176 pKa = 3.42PEE178 pKa = 4.72TKK180 pKa = 10.54NIAIPSSTEE189 pKa = 3.67VQEE192 pKa = 4.16WFPMEE197 pKa = 5.06DD198 pKa = 2.86VDD200 pKa = 5.71DD201 pKa = 4.46RR202 pKa = 11.84ARR204 pKa = 11.84EE205 pKa = 3.94PLDD208 pKa = 3.52GSEE211 pKa = 4.06THH213 pKa = 6.42QEE215 pKa = 3.64IAEE218 pKa = 4.39FACAMLPPGWCIVRR232 pKa = 11.84PEE234 pKa = 3.7GRR236 pKa = 11.84TYY238 pKa = 10.94ISTGPDD244 pKa = 2.96TSDD247 pKa = 3.37EE248 pKa = 4.45EE249 pKa = 4.19EE250 pKa = 4.7DD251 pKa = 3.41KK252 pKa = 11.57ALAPKK257 pKa = 9.88ILKK260 pKa = 6.96THH262 pKa = 6.76PSICPKK268 pKa = 10.21HH269 pKa = 5.88PRR271 pKa = 11.84HH272 pKa = 6.07HH273 pKa = 6.9KK274 pKa = 8.89IGGRR278 pKa = 11.84FVSDD282 pKa = 4.14LLDD285 pKa = 4.09SPSSSDD291 pKa = 3.59DD292 pKa = 3.58EE293 pKa = 4.47EE294 pKa = 4.07EE295 pKa = 4.12ARR297 pKa = 11.84QNLARR302 pKa = 11.84VLEE305 pKa = 4.37AALPHH310 pKa = 6.58PGEE313 pKa = 5.19DD314 pKa = 4.79SSDD317 pKa = 3.89DD318 pKa = 3.59DD319 pKa = 4.13PLYY322 pKa = 10.87LGEE325 pKa = 4.57PEE327 pKa = 4.27RR328 pKa = 11.84DD329 pKa = 3.25
MM1 pKa = 8.0RR2 pKa = 11.84NPGLGCLTWTGQLNGMGRR20 pKa = 11.84FQIQLFRR27 pKa = 11.84NLTLISVCLGSDD39 pKa = 3.19HH40 pKa = 7.15HH41 pKa = 8.38KK42 pKa = 11.12GITDD46 pKa = 3.65YY47 pKa = 11.41CCGPASCYY55 pKa = 9.72TIVWEE60 pKa = 4.65ISAEE64 pKa = 4.11VLATGTPYY72 pKa = 11.03LFRR75 pKa = 11.84AQPHH79 pKa = 5.94HH80 pKa = 7.2RR81 pKa = 11.84PLQTPAPWSKK91 pKa = 10.32RR92 pKa = 11.84LIRR95 pKa = 11.84TRR97 pKa = 11.84YY98 pKa = 8.45IRR100 pKa = 11.84NSVEE104 pKa = 3.66DD105 pKa = 3.71QFVHH109 pKa = 6.91FATEE113 pKa = 3.69ANYY116 pKa = 9.91TPLSAQALSIKK127 pKa = 10.11AIRR130 pKa = 11.84WHH132 pKa = 6.6KK133 pKa = 10.28GQLACEE139 pKa = 4.07GTPRR143 pKa = 11.84IGQAGIPEE151 pKa = 4.72TIYY154 pKa = 11.11AGIKK158 pKa = 9.37AYY160 pKa = 10.19GWFIPGKK167 pKa = 9.21KK168 pKa = 9.52EE169 pKa = 3.56RR170 pKa = 11.84VSWNEE175 pKa = 3.53DD176 pKa = 3.42PEE178 pKa = 4.72TKK180 pKa = 10.54NIAIPSSTEE189 pKa = 3.67VQEE192 pKa = 4.16WFPMEE197 pKa = 5.06DD198 pKa = 2.86VDD200 pKa = 5.71DD201 pKa = 4.46RR202 pKa = 11.84ARR204 pKa = 11.84EE205 pKa = 3.94PLDD208 pKa = 3.52GSEE211 pKa = 4.06THH213 pKa = 6.42QEE215 pKa = 3.64IAEE218 pKa = 4.39FACAMLPPGWCIVRR232 pKa = 11.84PEE234 pKa = 3.7GRR236 pKa = 11.84TYY238 pKa = 10.94ISTGPDD244 pKa = 2.96TSDD247 pKa = 3.37EE248 pKa = 4.45EE249 pKa = 4.19EE250 pKa = 4.7DD251 pKa = 3.41KK252 pKa = 11.57ALAPKK257 pKa = 9.88ILKK260 pKa = 6.96THH262 pKa = 6.76PSICPKK268 pKa = 10.21HH269 pKa = 5.88PRR271 pKa = 11.84HH272 pKa = 6.07HH273 pKa = 6.9KK274 pKa = 8.89IGGRR278 pKa = 11.84FVSDD282 pKa = 4.14LLDD285 pKa = 4.09SPSSSDD291 pKa = 3.59DD292 pKa = 3.58EE293 pKa = 4.47EE294 pKa = 4.07EE295 pKa = 4.12ARR297 pKa = 11.84QNLARR302 pKa = 11.84VLEE305 pKa = 4.37AALPHH310 pKa = 6.58PGEE313 pKa = 5.19DD314 pKa = 4.79SSDD317 pKa = 3.89DD318 pKa = 3.59DD319 pKa = 4.13PLYY322 pKa = 10.87LGEE325 pKa = 4.57PEE327 pKa = 4.27RR328 pKa = 11.84DD329 pKa = 3.25
Molecular weight: 36.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9J4C8|Q9J4C8_9RETR Gag polyprotein OS=Equine foamy virus OX=109270 GN=gag PE=4 SV=1
MM1 pKa = 7.27AQNEE5 pKa = 4.52TFDD8 pKa = 3.89PVALQGYY15 pKa = 8.38YY16 pKa = 9.8PAGGILADD24 pKa = 3.75NDD26 pKa = 3.79IINIRR31 pKa = 11.84FTSGQWGIGDD41 pKa = 3.31RR42 pKa = 11.84WLQVRR47 pKa = 11.84LRR49 pKa = 11.84LVDD52 pKa = 4.04PNTGQPLAQPEE63 pKa = 4.28YY64 pKa = 10.28EE65 pKa = 4.4DD66 pKa = 3.41TGLPAEE72 pKa = 4.1NRR74 pKa = 11.84GIVVAVSHH82 pKa = 5.89NAARR86 pKa = 11.84NIFNNVQPAGGPNRR100 pKa = 11.84HH101 pKa = 6.06GPLHH105 pKa = 7.19DD106 pKa = 4.03GQFQVGDD113 pKa = 3.89DD114 pKa = 3.96PSEE117 pKa = 4.08HH118 pKa = 6.0FVPIEE123 pKa = 3.93EE124 pKa = 4.01NLIPQEE130 pKa = 4.01IVNLGAARR138 pKa = 11.84RR139 pKa = 11.84EE140 pKa = 3.95VRR142 pKa = 11.84LLRR145 pKa = 11.84EE146 pKa = 3.64MCVRR150 pKa = 11.84LLHH153 pKa = 6.49VRR155 pKa = 11.84RR156 pKa = 11.84QMMGMGMPGAIQPQPPVGPLPAPAQPPIPGPPVPPPVPPPAPPAPVNPPVPPVQPIHH213 pKa = 6.55HH214 pKa = 6.89LPITHH219 pKa = 6.28IRR221 pKa = 11.84AVIGEE226 pKa = 4.45TPAQIRR232 pKa = 11.84DD233 pKa = 3.61VPLWLAQSIPALTGVYY249 pKa = 9.66PAMDD253 pKa = 4.51AGTLTRR259 pKa = 11.84LVNAITARR267 pKa = 11.84HH268 pKa = 6.28PGLALGMNEE277 pKa = 3.53AGSWHH282 pKa = 6.31EE283 pKa = 4.02AVHH286 pKa = 6.66LIWQRR291 pKa = 11.84TFGATALHH299 pKa = 6.47ALSDD303 pKa = 3.94VLKK306 pKa = 11.17GIAQRR311 pKa = 11.84NGVVMALEE319 pKa = 4.28MGLMFTNDD327 pKa = 3.42DD328 pKa = 3.68WDD330 pKa = 4.17LTWSVIRR337 pKa = 11.84RR338 pKa = 11.84CLPGQASVVTIQARR352 pKa = 11.84LDD354 pKa = 3.63ALPNNQARR362 pKa = 11.84IIQAGFIIRR371 pKa = 11.84EE372 pKa = 4.21VYY374 pKa = 9.82EE375 pKa = 4.1VLGLDD380 pKa = 3.54PLGRR384 pKa = 11.84PLNFPGGLTQRR395 pKa = 11.84DD396 pKa = 3.33TAVPVTRR403 pKa = 11.84GRR405 pKa = 11.84GRR407 pKa = 11.84GRR409 pKa = 11.84TGPRR413 pKa = 11.84RR414 pKa = 11.84GPVLPVSSNQRR425 pKa = 11.84RR426 pKa = 11.84QEE428 pKa = 4.13TAGGNQPQTQPQQQNTFSNQTNQRR452 pKa = 11.84GNQRR456 pKa = 11.84QWQNRR461 pKa = 11.84GTDD464 pKa = 3.12SQRR467 pKa = 11.84RR468 pKa = 11.84YY469 pKa = 9.78FFRR472 pKa = 11.84PRR474 pKa = 11.84PSQPQRR480 pKa = 11.84YY481 pKa = 8.87GSNQGPDD488 pKa = 3.08NPNPYY493 pKa = 9.72RR494 pKa = 11.84GRR496 pKa = 11.84DD497 pKa = 3.38STNQSGQEE505 pKa = 3.78RR506 pKa = 11.84QLPQQQQGSRR516 pKa = 11.84RR517 pKa = 11.84GPGRR521 pKa = 11.84NTNSGNNTVHH531 pKa = 6.51TVRR534 pKa = 11.84QVEE537 pKa = 4.45SSQLQQNASPTASPSTNQGQQPP559 pKa = 3.38
MM1 pKa = 7.27AQNEE5 pKa = 4.52TFDD8 pKa = 3.89PVALQGYY15 pKa = 8.38YY16 pKa = 9.8PAGGILADD24 pKa = 3.75NDD26 pKa = 3.79IINIRR31 pKa = 11.84FTSGQWGIGDD41 pKa = 3.31RR42 pKa = 11.84WLQVRR47 pKa = 11.84LRR49 pKa = 11.84LVDD52 pKa = 4.04PNTGQPLAQPEE63 pKa = 4.28YY64 pKa = 10.28EE65 pKa = 4.4DD66 pKa = 3.41TGLPAEE72 pKa = 4.1NRR74 pKa = 11.84GIVVAVSHH82 pKa = 5.89NAARR86 pKa = 11.84NIFNNVQPAGGPNRR100 pKa = 11.84HH101 pKa = 6.06GPLHH105 pKa = 7.19DD106 pKa = 4.03GQFQVGDD113 pKa = 3.89DD114 pKa = 3.96PSEE117 pKa = 4.08HH118 pKa = 6.0FVPIEE123 pKa = 3.93EE124 pKa = 4.01NLIPQEE130 pKa = 4.01IVNLGAARR138 pKa = 11.84RR139 pKa = 11.84EE140 pKa = 3.95VRR142 pKa = 11.84LLRR145 pKa = 11.84EE146 pKa = 3.64MCVRR150 pKa = 11.84LLHH153 pKa = 6.49VRR155 pKa = 11.84RR156 pKa = 11.84QMMGMGMPGAIQPQPPVGPLPAPAQPPIPGPPVPPPVPPPAPPAPVNPPVPPVQPIHH213 pKa = 6.55HH214 pKa = 6.89LPITHH219 pKa = 6.28IRR221 pKa = 11.84AVIGEE226 pKa = 4.45TPAQIRR232 pKa = 11.84DD233 pKa = 3.61VPLWLAQSIPALTGVYY249 pKa = 9.66PAMDD253 pKa = 4.51AGTLTRR259 pKa = 11.84LVNAITARR267 pKa = 11.84HH268 pKa = 6.28PGLALGMNEE277 pKa = 3.53AGSWHH282 pKa = 6.31EE283 pKa = 4.02AVHH286 pKa = 6.66LIWQRR291 pKa = 11.84TFGATALHH299 pKa = 6.47ALSDD303 pKa = 3.94VLKK306 pKa = 11.17GIAQRR311 pKa = 11.84NGVVMALEE319 pKa = 4.28MGLMFTNDD327 pKa = 3.42DD328 pKa = 3.68WDD330 pKa = 4.17LTWSVIRR337 pKa = 11.84RR338 pKa = 11.84CLPGQASVVTIQARR352 pKa = 11.84LDD354 pKa = 3.63ALPNNQARR362 pKa = 11.84IIQAGFIIRR371 pKa = 11.84EE372 pKa = 4.21VYY374 pKa = 9.82EE375 pKa = 4.1VLGLDD380 pKa = 3.54PLGRR384 pKa = 11.84PLNFPGGLTQRR395 pKa = 11.84DD396 pKa = 3.33TAVPVTRR403 pKa = 11.84GRR405 pKa = 11.84GRR407 pKa = 11.84GRR409 pKa = 11.84TGPRR413 pKa = 11.84RR414 pKa = 11.84GPVLPVSSNQRR425 pKa = 11.84RR426 pKa = 11.84QEE428 pKa = 4.13TAGGNQPQTQPQQQNTFSNQTNQRR452 pKa = 11.84GNQRR456 pKa = 11.84QWQNRR461 pKa = 11.84GTDD464 pKa = 3.12SQRR467 pKa = 11.84RR468 pKa = 11.84YY469 pKa = 9.78FFRR472 pKa = 11.84PRR474 pKa = 11.84PSQPQRR480 pKa = 11.84YY481 pKa = 8.87GSNQGPDD488 pKa = 3.08NPNPYY493 pKa = 9.72RR494 pKa = 11.84GRR496 pKa = 11.84DD497 pKa = 3.38STNQSGQEE505 pKa = 3.78RR506 pKa = 11.84QLPQQQQGSRR516 pKa = 11.84RR517 pKa = 11.84GPGRR521 pKa = 11.84NTNSGNNTVHH531 pKa = 6.51TVRR534 pKa = 11.84QVEE537 pKa = 4.45SSQLQQNASPTASPSTNQGQQPP559 pKa = 3.38
Molecular weight: 61.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3276 |
249 |
1153 |
655.2 |
74.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.739 ± 0.519 | 1.496 ± 0.425 |
5.189 ± 0.378 | 5.037 ± 0.468 |
2.564 ± 0.089 | 5.952 ± 0.908 |
2.717 ± 0.136 | 6.746 ± 0.651 |
5.464 ± 1.666 | 9.799 ± 0.744 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.832 ± 0.255 | 4.67 ± 0.398 |
7.54 ± 0.95 | 6.441 ± 0.687 |
5.464 ± 1.161 | 5.403 ± 0.583 |
6.441 ± 0.544 | 5.586 ± 0.459 |
2.106 ± 0.168 | 3.816 ± 0.636 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
