
Long Island tick rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sawgrhavirus; Island sawgrhavirus
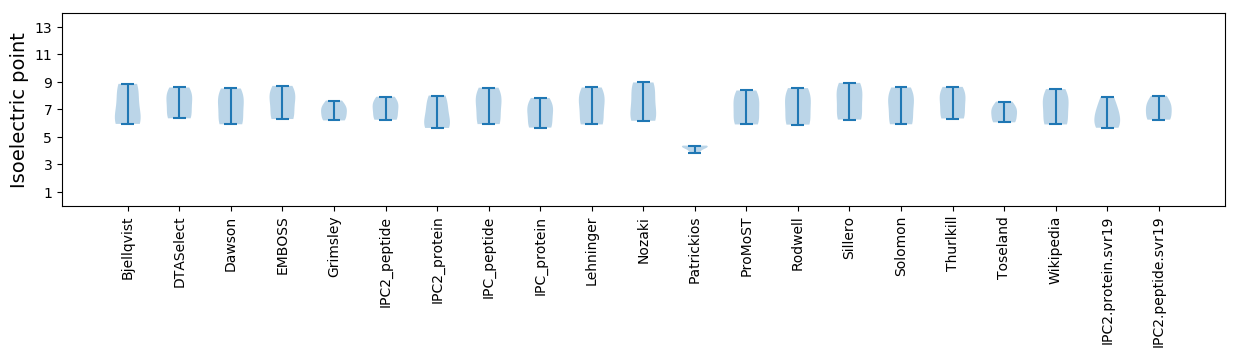
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|W8QQQ8|W8QQQ8_9RHAB GDP polyribonucleotidyltransferase OS=Long Island tick rhabdovirus OX=1459044 PE=4 SV=1
MM1 pKa = 7.5ASKK4 pKa = 10.62RR5 pKa = 11.84LDD7 pKa = 3.52VNLPNCDD14 pKa = 3.43KK15 pKa = 11.09KK16 pKa = 11.24SVNAGLPDD24 pKa = 3.56TKK26 pKa = 10.97VSVVYY31 pKa = 9.81PSEE34 pKa = 3.84WFKK37 pKa = 11.1TNEE40 pKa = 3.78GKK42 pKa = 10.65KK43 pKa = 9.89PIVRR47 pKa = 11.84IYY49 pKa = 10.46RR50 pKa = 11.84ATSTEE55 pKa = 4.56LYY57 pKa = 9.6RR58 pKa = 11.84ALYY61 pKa = 8.63PQIKK65 pKa = 9.41AEE67 pKa = 4.04KK68 pKa = 8.84WDD70 pKa = 4.06DD71 pKa = 3.65ALVTSFLVATLSEE84 pKa = 4.36RR85 pKa = 11.84PDD87 pKa = 3.55LFRR90 pKa = 11.84VEE92 pKa = 5.35SDD94 pKa = 3.78EE95 pKa = 4.03EE96 pKa = 4.31WVSFKK101 pKa = 10.76QQLSVGGQVSVTDD114 pKa = 4.28LLTLEE119 pKa = 4.33EE120 pKa = 5.08LEE122 pKa = 4.88AKK124 pKa = 10.27PPVPGPSKK132 pKa = 8.98PTPAEE137 pKa = 4.2EE138 pKa = 4.15ISFFCALLTIYY149 pKa = 10.44RR150 pKa = 11.84RR151 pKa = 11.84VQARR155 pKa = 11.84EE156 pKa = 3.76NPAVKK161 pKa = 10.47ASDD164 pKa = 3.55LTDD167 pKa = 2.99KK168 pKa = 10.85FKK170 pKa = 11.36NVLGRR175 pKa = 11.84EE176 pKa = 4.33PYY178 pKa = 11.0NMSDD182 pKa = 2.98EE183 pKa = 4.65DD184 pKa = 3.9ARR186 pKa = 11.84LMVPIAGTANLTSGFMRR203 pKa = 11.84ILSVIDD209 pKa = 3.17MFFVKK214 pKa = 10.26FPQAPGSAVRR224 pKa = 11.84IATFTLRR231 pKa = 11.84YY232 pKa = 9.05RR233 pKa = 11.84GCTAITALTHH243 pKa = 6.21ACITLGLDD251 pKa = 3.52AYY253 pKa = 10.47DD254 pKa = 4.07LASLIVPQSPADD266 pKa = 3.63DD267 pKa = 3.4AVRR270 pKa = 11.84VYY272 pKa = 11.34EE273 pKa = 5.41DD274 pKa = 3.88GQEE277 pKa = 3.96HH278 pKa = 7.79DD279 pKa = 4.63DD280 pKa = 3.99PYY282 pKa = 11.67GYY284 pKa = 10.41FPYY287 pKa = 10.02HH288 pKa = 5.94FVMGLSNHH296 pKa = 5.74SPYY299 pKa = 11.17SLAAAPSLTFYY310 pKa = 9.64TRR312 pKa = 11.84CLAAFGGSKK321 pKa = 9.69NAAYY325 pKa = 8.53TYY327 pKa = 8.81TAAGIRR333 pKa = 11.84NVPEE337 pKa = 4.86LIEE340 pKa = 3.85TAYY343 pKa = 11.39LLIKK347 pKa = 9.96KK348 pKa = 9.41IRR350 pKa = 11.84RR351 pKa = 11.84LEE353 pKa = 4.28PGDD356 pKa = 3.86LRR358 pKa = 11.84FAEE361 pKa = 4.59SSAMDD366 pKa = 2.9KK367 pKa = 10.51WKK369 pKa = 10.43AHH371 pKa = 5.95KK372 pKa = 9.54EE373 pKa = 4.01ARR375 pKa = 11.84DD376 pKa = 3.76KK377 pKa = 11.24LLKK380 pKa = 10.64DD381 pKa = 3.71DD382 pKa = 6.22DD383 pKa = 4.28EE384 pKa = 7.18DD385 pKa = 3.82EE386 pKa = 4.53VQLHH390 pKa = 6.47EE391 pKa = 4.61GAQEE395 pKa = 4.05RR396 pKa = 11.84GLVASLLALPTRR408 pKa = 11.84QEE410 pKa = 3.88LTPLHH415 pKa = 6.3RR416 pKa = 11.84RR417 pKa = 11.84IDD419 pKa = 3.53EE420 pKa = 4.38ALAEE424 pKa = 4.28LSVHH428 pKa = 6.53HH429 pKa = 6.26GLSRR433 pKa = 11.84KK434 pKa = 9.04NAMGLYY440 pKa = 9.94HH441 pKa = 7.72AAQCFKK447 pKa = 10.58GADD450 pKa = 3.53VGTVGHH456 pKa = 7.06TIYY459 pKa = 10.57DD460 pKa = 3.34QYY462 pKa = 11.68GRR464 pKa = 11.84VPPYY468 pKa = 10.84VEE470 pKa = 3.8
MM1 pKa = 7.5ASKK4 pKa = 10.62RR5 pKa = 11.84LDD7 pKa = 3.52VNLPNCDD14 pKa = 3.43KK15 pKa = 11.09KK16 pKa = 11.24SVNAGLPDD24 pKa = 3.56TKK26 pKa = 10.97VSVVYY31 pKa = 9.81PSEE34 pKa = 3.84WFKK37 pKa = 11.1TNEE40 pKa = 3.78GKK42 pKa = 10.65KK43 pKa = 9.89PIVRR47 pKa = 11.84IYY49 pKa = 10.46RR50 pKa = 11.84ATSTEE55 pKa = 4.56LYY57 pKa = 9.6RR58 pKa = 11.84ALYY61 pKa = 8.63PQIKK65 pKa = 9.41AEE67 pKa = 4.04KK68 pKa = 8.84WDD70 pKa = 4.06DD71 pKa = 3.65ALVTSFLVATLSEE84 pKa = 4.36RR85 pKa = 11.84PDD87 pKa = 3.55LFRR90 pKa = 11.84VEE92 pKa = 5.35SDD94 pKa = 3.78EE95 pKa = 4.03EE96 pKa = 4.31WVSFKK101 pKa = 10.76QQLSVGGQVSVTDD114 pKa = 4.28LLTLEE119 pKa = 4.33EE120 pKa = 5.08LEE122 pKa = 4.88AKK124 pKa = 10.27PPVPGPSKK132 pKa = 8.98PTPAEE137 pKa = 4.2EE138 pKa = 4.15ISFFCALLTIYY149 pKa = 10.44RR150 pKa = 11.84RR151 pKa = 11.84VQARR155 pKa = 11.84EE156 pKa = 3.76NPAVKK161 pKa = 10.47ASDD164 pKa = 3.55LTDD167 pKa = 2.99KK168 pKa = 10.85FKK170 pKa = 11.36NVLGRR175 pKa = 11.84EE176 pKa = 4.33PYY178 pKa = 11.0NMSDD182 pKa = 2.98EE183 pKa = 4.65DD184 pKa = 3.9ARR186 pKa = 11.84LMVPIAGTANLTSGFMRR203 pKa = 11.84ILSVIDD209 pKa = 3.17MFFVKK214 pKa = 10.26FPQAPGSAVRR224 pKa = 11.84IATFTLRR231 pKa = 11.84YY232 pKa = 9.05RR233 pKa = 11.84GCTAITALTHH243 pKa = 6.21ACITLGLDD251 pKa = 3.52AYY253 pKa = 10.47DD254 pKa = 4.07LASLIVPQSPADD266 pKa = 3.63DD267 pKa = 3.4AVRR270 pKa = 11.84VYY272 pKa = 11.34EE273 pKa = 5.41DD274 pKa = 3.88GQEE277 pKa = 3.96HH278 pKa = 7.79DD279 pKa = 4.63DD280 pKa = 3.99PYY282 pKa = 11.67GYY284 pKa = 10.41FPYY287 pKa = 10.02HH288 pKa = 5.94FVMGLSNHH296 pKa = 5.74SPYY299 pKa = 11.17SLAAAPSLTFYY310 pKa = 9.64TRR312 pKa = 11.84CLAAFGGSKK321 pKa = 9.69NAAYY325 pKa = 8.53TYY327 pKa = 8.81TAAGIRR333 pKa = 11.84NVPEE337 pKa = 4.86LIEE340 pKa = 3.85TAYY343 pKa = 11.39LLIKK347 pKa = 9.96KK348 pKa = 9.41IRR350 pKa = 11.84RR351 pKa = 11.84LEE353 pKa = 4.28PGDD356 pKa = 3.86LRR358 pKa = 11.84FAEE361 pKa = 4.59SSAMDD366 pKa = 2.9KK367 pKa = 10.51WKK369 pKa = 10.43AHH371 pKa = 5.95KK372 pKa = 9.54EE373 pKa = 4.01ARR375 pKa = 11.84DD376 pKa = 3.76KK377 pKa = 11.24LLKK380 pKa = 10.64DD381 pKa = 3.71DD382 pKa = 6.22DD383 pKa = 4.28EE384 pKa = 7.18DD385 pKa = 3.82EE386 pKa = 4.53VQLHH390 pKa = 6.47EE391 pKa = 4.61GAQEE395 pKa = 4.05RR396 pKa = 11.84GLVASLLALPTRR408 pKa = 11.84QEE410 pKa = 3.88LTPLHH415 pKa = 6.3RR416 pKa = 11.84RR417 pKa = 11.84IDD419 pKa = 3.53EE420 pKa = 4.38ALAEE424 pKa = 4.28LSVHH428 pKa = 6.53HH429 pKa = 6.26GLSRR433 pKa = 11.84KK434 pKa = 9.04NAMGLYY440 pKa = 9.94HH441 pKa = 7.72AAQCFKK447 pKa = 10.58GADD450 pKa = 3.53VGTVGHH456 pKa = 7.06TIYY459 pKa = 10.57DD460 pKa = 3.34QYY462 pKa = 11.68GRR464 pKa = 11.84VPPYY468 pKa = 10.84VEE470 pKa = 3.8
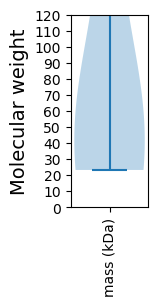
Molecular weight: 52.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8R902|W8R902_9RHAB Glycoprotein OS=Long Island tick rhabdovirus OX=1459044 PE=3 SV=1
MM1 pKa = 7.26LRR3 pKa = 11.84YY4 pKa = 10.05AFVLLMIAVAFGWEE18 pKa = 4.22PEE20 pKa = 4.12LGEE23 pKa = 4.18YY24 pKa = 8.21WVPSPISPWRR34 pKa = 11.84LATKK38 pKa = 10.07TDD40 pKa = 4.13FTCPSIRR47 pKa = 11.84VDD49 pKa = 3.29PVSPLKK55 pKa = 10.67AINEE59 pKa = 4.51SYY61 pKa = 11.06VEE63 pKa = 4.11YY64 pKa = 8.64PTMGGARR71 pKa = 11.84KK72 pKa = 7.97EE73 pKa = 4.2HH74 pKa = 6.71PGWLCIRR81 pKa = 11.84KK82 pKa = 7.75TYY84 pKa = 8.73QTTCDD89 pKa = 3.34TNFWGHH95 pKa = 4.54QTIKK99 pKa = 10.46HH100 pKa = 5.42EE101 pKa = 4.19EE102 pKa = 3.75WSIVADD108 pKa = 3.98VEE110 pKa = 4.24EE111 pKa = 4.53CRR113 pKa = 11.84QAVSHH118 pKa = 5.75YY119 pKa = 10.7QLGTYY124 pKa = 8.77EE125 pKa = 4.83DD126 pKa = 4.42PRR128 pKa = 11.84HH129 pKa = 6.49PDD131 pKa = 3.59PVCTWMAISSTYY143 pKa = 10.25RR144 pKa = 11.84AGTLLLPHH152 pKa = 6.32TTLVDD157 pKa = 3.53PFSYY161 pKa = 10.43TFVDD165 pKa = 3.57SLFPGTHH172 pKa = 6.88CKK174 pKa = 9.73TIPCMTVHH182 pKa = 7.51PDD184 pKa = 3.81VLWHH188 pKa = 6.28SSSTITEE195 pKa = 4.22NCPMANGIHH204 pKa = 6.1IKK206 pKa = 10.43LYY208 pKa = 9.5NDD210 pKa = 2.47NRR212 pKa = 11.84ARR214 pKa = 11.84KK215 pKa = 8.67KK216 pKa = 8.68AHH218 pKa = 6.05EE219 pKa = 4.21WLSVNDD225 pKa = 4.57GPLIPLGGSCSMHH238 pKa = 6.05YY239 pKa = 10.17CGQAGLRR246 pKa = 11.84TPGGIWYY253 pKa = 8.4PYY255 pKa = 10.23LGRR258 pKa = 11.84QKK260 pKa = 10.9YY261 pKa = 8.78PEE263 pKa = 4.61CKK265 pKa = 9.18EE266 pKa = 3.65AWRR269 pKa = 11.84ATPTPAGNGAKK280 pKa = 10.03LDD282 pKa = 3.71IEE284 pKa = 4.24LQEE287 pKa = 4.3LIARR291 pKa = 11.84RR292 pKa = 11.84EE293 pKa = 4.16CLNVLQQIRR302 pKa = 11.84SGVSPTFHH310 pKa = 6.62MLSHH314 pKa = 6.37FQPRR318 pKa = 11.84HH319 pKa = 4.21TGYY322 pKa = 10.6YY323 pKa = 8.77RR324 pKa = 11.84VYY326 pKa = 10.75RR327 pKa = 11.84MNNGLVEE334 pKa = 4.15YY335 pKa = 10.39SLALYY340 pKa = 10.54APIFNITMPPSINFGVRR357 pKa = 11.84RR358 pKa = 11.84NKK360 pKa = 10.07SRR362 pKa = 11.84YY363 pKa = 7.6HH364 pKa = 7.58LPMIEE369 pKa = 4.16SGHH372 pKa = 6.13PGVWSAFNGIHH383 pKa = 6.21IMHH386 pKa = 7.24NKK388 pKa = 7.72TVIVPEE394 pKa = 3.99LEE396 pKa = 4.15LYY398 pKa = 10.01KK399 pKa = 10.46EE400 pKa = 4.75HH401 pKa = 7.38YY402 pKa = 10.33SEE404 pKa = 4.02TLFYY408 pKa = 11.24YY409 pKa = 10.41KK410 pKa = 10.49AQLAEE415 pKa = 4.47HH416 pKa = 7.26PSQVRR421 pKa = 11.84QANSTDD427 pKa = 3.12ITPHH431 pKa = 5.71TKK433 pKa = 8.11TTVTVKK439 pKa = 10.63RR440 pKa = 11.84PTLKK444 pKa = 10.58AWFSTMWDD452 pKa = 3.82SLWGKK457 pKa = 9.15VVSITGIILAIFTTGFLIKK476 pKa = 10.16VLPWSRR482 pKa = 11.84LFRR485 pKa = 11.84RR486 pKa = 11.84PQPMHH491 pKa = 6.12PQIVHH496 pKa = 4.89YY497 pKa = 9.21TPATGSVSWW506 pKa = 3.73
MM1 pKa = 7.26LRR3 pKa = 11.84YY4 pKa = 10.05AFVLLMIAVAFGWEE18 pKa = 4.22PEE20 pKa = 4.12LGEE23 pKa = 4.18YY24 pKa = 8.21WVPSPISPWRR34 pKa = 11.84LATKK38 pKa = 10.07TDD40 pKa = 4.13FTCPSIRR47 pKa = 11.84VDD49 pKa = 3.29PVSPLKK55 pKa = 10.67AINEE59 pKa = 4.51SYY61 pKa = 11.06VEE63 pKa = 4.11YY64 pKa = 8.64PTMGGARR71 pKa = 11.84KK72 pKa = 7.97EE73 pKa = 4.2HH74 pKa = 6.71PGWLCIRR81 pKa = 11.84KK82 pKa = 7.75TYY84 pKa = 8.73QTTCDD89 pKa = 3.34TNFWGHH95 pKa = 4.54QTIKK99 pKa = 10.46HH100 pKa = 5.42EE101 pKa = 4.19EE102 pKa = 3.75WSIVADD108 pKa = 3.98VEE110 pKa = 4.24EE111 pKa = 4.53CRR113 pKa = 11.84QAVSHH118 pKa = 5.75YY119 pKa = 10.7QLGTYY124 pKa = 8.77EE125 pKa = 4.83DD126 pKa = 4.42PRR128 pKa = 11.84HH129 pKa = 6.49PDD131 pKa = 3.59PVCTWMAISSTYY143 pKa = 10.25RR144 pKa = 11.84AGTLLLPHH152 pKa = 6.32TTLVDD157 pKa = 3.53PFSYY161 pKa = 10.43TFVDD165 pKa = 3.57SLFPGTHH172 pKa = 6.88CKK174 pKa = 9.73TIPCMTVHH182 pKa = 7.51PDD184 pKa = 3.81VLWHH188 pKa = 6.28SSSTITEE195 pKa = 4.22NCPMANGIHH204 pKa = 6.1IKK206 pKa = 10.43LYY208 pKa = 9.5NDD210 pKa = 2.47NRR212 pKa = 11.84ARR214 pKa = 11.84KK215 pKa = 8.67KK216 pKa = 8.68AHH218 pKa = 6.05EE219 pKa = 4.21WLSVNDD225 pKa = 4.57GPLIPLGGSCSMHH238 pKa = 6.05YY239 pKa = 10.17CGQAGLRR246 pKa = 11.84TPGGIWYY253 pKa = 8.4PYY255 pKa = 10.23LGRR258 pKa = 11.84QKK260 pKa = 10.9YY261 pKa = 8.78PEE263 pKa = 4.61CKK265 pKa = 9.18EE266 pKa = 3.65AWRR269 pKa = 11.84ATPTPAGNGAKK280 pKa = 10.03LDD282 pKa = 3.71IEE284 pKa = 4.24LQEE287 pKa = 4.3LIARR291 pKa = 11.84RR292 pKa = 11.84EE293 pKa = 4.16CLNVLQQIRR302 pKa = 11.84SGVSPTFHH310 pKa = 6.62MLSHH314 pKa = 6.37FQPRR318 pKa = 11.84HH319 pKa = 4.21TGYY322 pKa = 10.6YY323 pKa = 8.77RR324 pKa = 11.84VYY326 pKa = 10.75RR327 pKa = 11.84MNNGLVEE334 pKa = 4.15YY335 pKa = 10.39SLALYY340 pKa = 10.54APIFNITMPPSINFGVRR357 pKa = 11.84RR358 pKa = 11.84NKK360 pKa = 10.07SRR362 pKa = 11.84YY363 pKa = 7.6HH364 pKa = 7.58LPMIEE369 pKa = 4.16SGHH372 pKa = 6.13PGVWSAFNGIHH383 pKa = 6.21IMHH386 pKa = 7.24NKK388 pKa = 7.72TVIVPEE394 pKa = 3.99LEE396 pKa = 4.15LYY398 pKa = 10.01KK399 pKa = 10.46EE400 pKa = 4.75HH401 pKa = 7.38YY402 pKa = 10.33SEE404 pKa = 4.02TLFYY408 pKa = 11.24YY409 pKa = 10.41KK410 pKa = 10.49AQLAEE415 pKa = 4.47HH416 pKa = 7.26PSQVRR421 pKa = 11.84QANSTDD427 pKa = 3.12ITPHH431 pKa = 5.71TKK433 pKa = 8.11TTVTVKK439 pKa = 10.63RR440 pKa = 11.84PTLKK444 pKa = 10.58AWFSTMWDD452 pKa = 3.82SLWGKK457 pKa = 9.15VVSITGIILAIFTTGFLIKK476 pKa = 10.16VLPWSRR482 pKa = 11.84LFRR485 pKa = 11.84RR486 pKa = 11.84PQPMHH491 pKa = 6.12PQIVHH496 pKa = 4.89YY497 pKa = 9.21TPATGSVSWW506 pKa = 3.73
Molecular weight: 57.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3588 |
205 |
2131 |
717.6 |
81.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.048 ± 0.897 | 1.895 ± 0.218 |
5.295 ± 0.567 | 6.048 ± 0.316 |
3.484 ± 0.35 | 6.159 ± 0.225 |
2.982 ± 0.451 | 4.849 ± 0.405 |
5.072 ± 0.294 | 10.117 ± 0.858 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.341 ± 0.201 | 3.372 ± 0.384 |
6.187 ± 1.057 | 3.122 ± 0.135 |
6.494 ± 0.346 | 7.079 ± 0.241 |
6.745 ± 0.457 | 7.023 ± 0.547 |
2.09 ± 0.35 | 3.595 ± 0.404 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
