
Pandoravirus quercus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified dsDNA viruses; Pandoravirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 1179 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
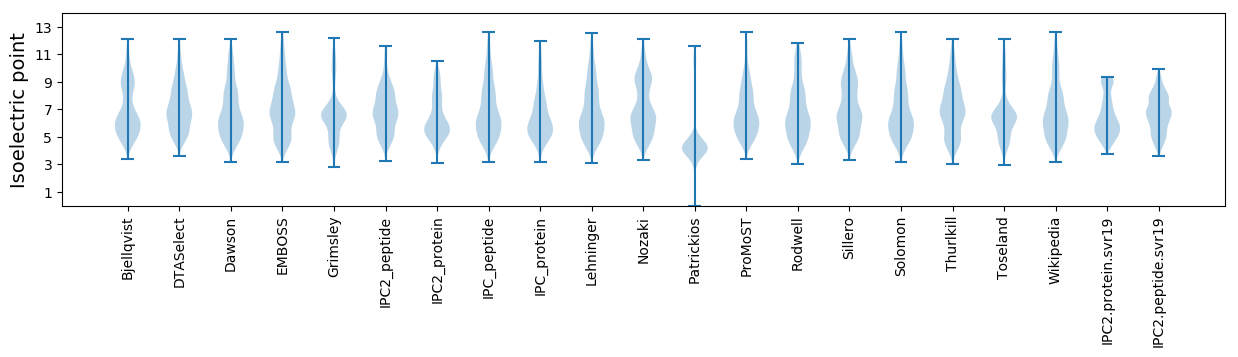
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U7UB16|A0A2U7UB16_9VIRU Uncharacterized protein OS=Pandoravirus quercus OX=2107709 GN=pqer_cds_1170 PE=4 SV=1
MM1 pKa = 7.67ARR3 pKa = 11.84PTTDD7 pKa = 2.13AWVYY11 pKa = 10.62EE12 pKa = 4.5DD13 pKa = 4.69GWPNDD18 pKa = 3.53DD19 pKa = 5.68DD20 pKa = 3.93EE21 pKa = 5.42TYY23 pKa = 10.77QCSDD27 pKa = 4.9DD28 pKa = 3.93NNQLAGWGGLHH39 pKa = 6.64GWDD42 pKa = 3.34DD43 pKa = 4.53CIYY46 pKa = 10.95NDD48 pKa = 3.06ILAYY52 pKa = 7.16QTRR55 pKa = 11.84DD56 pKa = 3.31DD57 pKa = 3.83TVYY60 pKa = 11.03NAIGSCIDD68 pKa = 4.11HH69 pKa = 6.78NNADD73 pKa = 4.1DD74 pKa = 4.23GTWGNGDD81 pKa = 4.42DD82 pKa = 4.44DD83 pKa = 4.69VSGLGEE89 pKa = 4.3CPVAPKK95 pKa = 8.61HH96 pKa = 5.17TPYY99 pKa = 10.9AYY101 pKa = 9.97GYY103 pKa = 7.53TFIEE107 pKa = 4.79GFMPADD113 pKa = 3.45SFDD116 pKa = 3.74ALVRR120 pKa = 11.84PEE122 pKa = 3.77RR123 pKa = 11.84RR124 pKa = 11.84LVVVVGGVPVALDD137 pKa = 3.27AAEE140 pKa = 4.13VEE142 pKa = 4.49RR143 pKa = 11.84AGNVGRR149 pKa = 11.84PLATPLGSCSLGPLDD164 pKa = 4.57VEE166 pKa = 4.62RR167 pKa = 11.84AHH169 pKa = 6.64EE170 pKa = 4.25ADD172 pKa = 3.09EE173 pKa = 4.16ALYY176 pKa = 10.4EE177 pKa = 4.15RR178 pKa = 11.84EE179 pKa = 3.78GLRR182 pKa = 11.84RR183 pKa = 11.84FLTTAPVTATIEE195 pKa = 3.94AAAPALGAGAGAAADD210 pKa = 3.52IAGGSILAGVLGGLFAALLL229 pKa = 4.05
MM1 pKa = 7.67ARR3 pKa = 11.84PTTDD7 pKa = 2.13AWVYY11 pKa = 10.62EE12 pKa = 4.5DD13 pKa = 4.69GWPNDD18 pKa = 3.53DD19 pKa = 5.68DD20 pKa = 3.93EE21 pKa = 5.42TYY23 pKa = 10.77QCSDD27 pKa = 4.9DD28 pKa = 3.93NNQLAGWGGLHH39 pKa = 6.64GWDD42 pKa = 3.34DD43 pKa = 4.53CIYY46 pKa = 10.95NDD48 pKa = 3.06ILAYY52 pKa = 7.16QTRR55 pKa = 11.84DD56 pKa = 3.31DD57 pKa = 3.83TVYY60 pKa = 11.03NAIGSCIDD68 pKa = 4.11HH69 pKa = 6.78NNADD73 pKa = 4.1DD74 pKa = 4.23GTWGNGDD81 pKa = 4.42DD82 pKa = 4.44DD83 pKa = 4.69VSGLGEE89 pKa = 4.3CPVAPKK95 pKa = 8.61HH96 pKa = 5.17TPYY99 pKa = 10.9AYY101 pKa = 9.97GYY103 pKa = 7.53TFIEE107 pKa = 4.79GFMPADD113 pKa = 3.45SFDD116 pKa = 3.74ALVRR120 pKa = 11.84PEE122 pKa = 3.77RR123 pKa = 11.84RR124 pKa = 11.84LVVVVGGVPVALDD137 pKa = 3.27AAEE140 pKa = 4.13VEE142 pKa = 4.49RR143 pKa = 11.84AGNVGRR149 pKa = 11.84PLATPLGSCSLGPLDD164 pKa = 4.57VEE166 pKa = 4.62RR167 pKa = 11.84AHH169 pKa = 6.64EE170 pKa = 4.25ADD172 pKa = 3.09EE173 pKa = 4.16ALYY176 pKa = 10.4EE177 pKa = 4.15RR178 pKa = 11.84EE179 pKa = 3.78GLRR182 pKa = 11.84RR183 pKa = 11.84FLTTAPVTATIEE195 pKa = 3.94AAAPALGAGAGAAADD210 pKa = 3.52IAGGSILAGVLGGLFAALLL229 pKa = 4.05
Molecular weight: 24.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U7U9V9|A0A2U7U9V9_9VIRU Uncharacterized protein OS=Pandoravirus quercus OX=2107709 GN=pqer_cds_794 PE=4 SV=1
MM1 pKa = 7.71DD2 pKa = 5.2TLPAEE7 pKa = 4.63IGCMILGYY15 pKa = 10.28LEE17 pKa = 4.38PTWLASVSMASRR29 pKa = 11.84GLYY32 pKa = 9.1HH33 pKa = 7.17WGALVCRR40 pKa = 11.84GEE42 pKa = 4.27ALRR45 pKa = 11.84MPRR48 pKa = 11.84NAMEE52 pKa = 3.79VAAANGAWDD61 pKa = 4.16FMQWLRR67 pKa = 11.84HH68 pKa = 5.03HH69 pKa = 7.32LYY71 pKa = 10.01WPWTPVTLATAAVHH85 pKa = 5.24NQRR88 pKa = 11.84LVFKK92 pKa = 10.22RR93 pKa = 11.84LFKK96 pKa = 11.05GRR98 pKa = 11.84TCSVDD103 pKa = 2.95ARR105 pKa = 11.84AAAAALVGGGPALLCTALRR124 pKa = 11.84HH125 pKa = 5.49GCPKK129 pKa = 10.47SGLLTTVACLLGRR142 pKa = 11.84DD143 pKa = 4.01GLASRR148 pKa = 11.84LMRR151 pKa = 11.84TTVPDD156 pKa = 4.36DD157 pKa = 3.78LMNLCGLATRR167 pKa = 11.84NWQILSFMTPGDD179 pKa = 3.22WRR181 pKa = 11.84RR182 pKa = 11.84TRR184 pKa = 11.84RR185 pKa = 11.84WLSDD189 pKa = 3.49PDD191 pKa = 3.47RR192 pKa = 11.84APIDD196 pKa = 3.66LRR198 pKa = 11.84VAAHH202 pKa = 5.83QLAEE206 pKa = 4.38HH207 pKa = 7.26RR208 pKa = 11.84DD209 pKa = 3.87TLAGLAKK216 pKa = 10.22KK217 pKa = 10.09LALGEE222 pKa = 4.07IARR225 pKa = 11.84RR226 pKa = 11.84PPPPPPPRR234 pKa = 11.84LPLGAGYY241 pKa = 9.38PDD243 pKa = 3.48WLHH246 pKa = 6.0YY247 pKa = 10.16CIDD250 pKa = 3.87MIRR253 pKa = 11.84WKK255 pKa = 10.43NDD257 pKa = 2.78CLFRR261 pKa = 11.84EE262 pKa = 4.11RR263 pKa = 11.84RR264 pKa = 11.84QWHH267 pKa = 5.91EE268 pKa = 4.11EE269 pKa = 4.05SPQRR273 pKa = 11.84WPKK276 pKa = 10.01PVLADD281 pKa = 4.35RR282 pKa = 11.84SPGPEE287 pKa = 4.14SKK289 pKa = 10.58CRR291 pKa = 11.84TPVEE295 pKa = 3.77KK296 pKa = 10.5RR297 pKa = 11.84QPHH300 pKa = 6.24TIARR304 pKa = 11.84MQPRR308 pKa = 11.84RR309 pKa = 11.84HH310 pKa = 6.01ARR312 pKa = 11.84RR313 pKa = 11.84PWRR316 pKa = 3.84
MM1 pKa = 7.71DD2 pKa = 5.2TLPAEE7 pKa = 4.63IGCMILGYY15 pKa = 10.28LEE17 pKa = 4.38PTWLASVSMASRR29 pKa = 11.84GLYY32 pKa = 9.1HH33 pKa = 7.17WGALVCRR40 pKa = 11.84GEE42 pKa = 4.27ALRR45 pKa = 11.84MPRR48 pKa = 11.84NAMEE52 pKa = 3.79VAAANGAWDD61 pKa = 4.16FMQWLRR67 pKa = 11.84HH68 pKa = 5.03HH69 pKa = 7.32LYY71 pKa = 10.01WPWTPVTLATAAVHH85 pKa = 5.24NQRR88 pKa = 11.84LVFKK92 pKa = 10.22RR93 pKa = 11.84LFKK96 pKa = 11.05GRR98 pKa = 11.84TCSVDD103 pKa = 2.95ARR105 pKa = 11.84AAAAALVGGGPALLCTALRR124 pKa = 11.84HH125 pKa = 5.49GCPKK129 pKa = 10.47SGLLTTVACLLGRR142 pKa = 11.84DD143 pKa = 4.01GLASRR148 pKa = 11.84LMRR151 pKa = 11.84TTVPDD156 pKa = 4.36DD157 pKa = 3.78LMNLCGLATRR167 pKa = 11.84NWQILSFMTPGDD179 pKa = 3.22WRR181 pKa = 11.84RR182 pKa = 11.84TRR184 pKa = 11.84RR185 pKa = 11.84WLSDD189 pKa = 3.49PDD191 pKa = 3.47RR192 pKa = 11.84APIDD196 pKa = 3.66LRR198 pKa = 11.84VAAHH202 pKa = 5.83QLAEE206 pKa = 4.38HH207 pKa = 7.26RR208 pKa = 11.84DD209 pKa = 3.87TLAGLAKK216 pKa = 10.22KK217 pKa = 10.09LALGEE222 pKa = 4.07IARR225 pKa = 11.84RR226 pKa = 11.84PPPPPPPRR234 pKa = 11.84LPLGAGYY241 pKa = 9.38PDD243 pKa = 3.48WLHH246 pKa = 6.0YY247 pKa = 10.16CIDD250 pKa = 3.87MIRR253 pKa = 11.84WKK255 pKa = 10.43NDD257 pKa = 2.78CLFRR261 pKa = 11.84EE262 pKa = 4.11RR263 pKa = 11.84RR264 pKa = 11.84QWHH267 pKa = 5.91EE268 pKa = 4.11EE269 pKa = 4.05SPQRR273 pKa = 11.84WPKK276 pKa = 10.01PVLADD281 pKa = 4.35RR282 pKa = 11.84SPGPEE287 pKa = 4.14SKK289 pKa = 10.58CRR291 pKa = 11.84TPVEE295 pKa = 3.77KK296 pKa = 10.5RR297 pKa = 11.84QPHH300 pKa = 6.24TIARR304 pKa = 11.84MQPRR308 pKa = 11.84RR309 pKa = 11.84HH310 pKa = 6.01ARR312 pKa = 11.84RR313 pKa = 11.84PWRR316 pKa = 3.84
Molecular weight: 35.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
468644 |
57 |
2278 |
397.5 |
42.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.946 ± 0.082 | 2.536 ± 0.048 |
7.561 ± 0.078 | 3.831 ± 0.051 |
2.22 ± 0.027 | 7.937 ± 0.078 |
3.01 ± 0.05 | 3.223 ± 0.031 |
2.01 ± 0.048 | 8.122 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.079 ± 0.025 | 2.241 ± 0.039 |
6.645 ± 0.06 | 2.74 ± 0.043 |
8.472 ± 0.068 | 5.842 ± 0.063 |
6.009 ± 0.059 | 7.388 ± 0.051 |
2.029 ± 0.037 | 2.16 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
